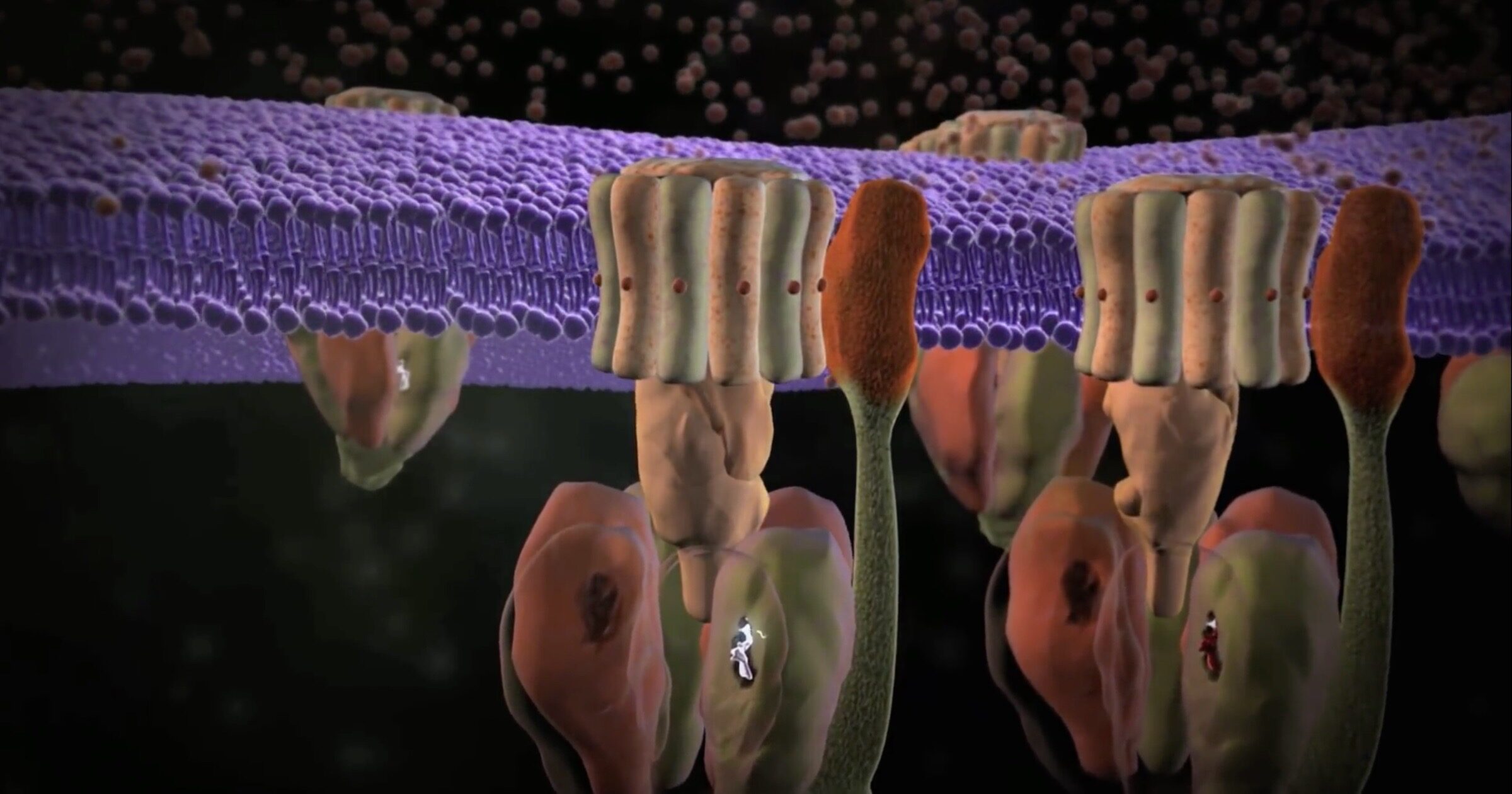
ATP Synthase
In 1997, John E. Walker shared the Nobel Prize in Chemistry with Paul Boyer and Jens Skou for discovering that ATP was synthesized in cells by a rotary engine. He was 56 at the time; now, at age 79, he is still engaged in research on the mechanism of ATP synthase (see our animation). Co-authoring a paper in PNAS,1 Walker and two others used cryo-electron microscopy to examine the motor at higher resolution than was possible 23 years ago. The abstract of their paper introduces news about this rotating marvel:
A "Grotthus mechanism" is a hand-off series something like a bucket brigade. The protons that power the rotor do not just flow by themselves; they are passed along to the inlet by a bucket brigade of water molecules, which take in each proton and hand it off to the next water molecule.The structure of the dimeric ATP synthase from bovine mitochondria determined in three rotational states by electron cryo-microscopy provides evidence that the proton uptake from the mitochondrial matrix via the proton inlet half channel proceeds via a Grotthus mechanism, and a similar mechanism may operate in the exit half channel. The structure has given information about the architecture and mechanical constitution and properties of the peripheral stalk, part of the membrane extrinsic region of the stator, and how the action of the peripheral stalk damps the side-to-side rocking motions that occur in the enzyme complex during the catalytic cycle. [Emphasis added.]
Several other interesting facts came to light in the paper. Like they say, there is a "side-to-side rocking motion" in the machine. This motor really hums! As the F1 portion rapidly churns out ATP molecules in a three-stage process, some rocking is inevitable. The peripheral stalk helps damp those motions. This helps to "increase the efficiency of the enzyme" by keeping the a-subunit in place — that is the subunit connected to the rotating Fo carousel in the "membrane domain" where Fo is anchored. A good designer would not want the rotor shaken apart by vibrations. The peripheral stalk takes care of that —
Synthase SystemsSome degree of motion of the catalytic domain must be permitted to allow efficient conversion of the rotational torque of the central stalk into conformational changes in the catalytic subunits, but without wasting energy by unneeded displacement of the extrinsic portion of the enzyme. The flexibility of the peripheral stalk effectively meters this balance, preventing free rotation of the catalytic domain, accommodating the rocking motions induced by the asymmetric central stalk, and damping displacement of the membrane domain helping to keep the a-subunit in place.
Since 1997, researchers also found that ATP synthase machines come in pairs (dimers). The dimers, further, are mounted in rows on folds of the cristae (the membranes within mitochondria). The precise angle between the two dimerized motors and the spacing between them maximizes proton intake. The new paper says that the angle between the dimers flexes a bit, damping vibrations even more, thanks to specific molecules at the pivot of the wedge that allow some flexing, but not too much:
Scientists at Austria's Institute of Science and Technology have also been working on ATP synthase. They boast that they have solved the structure of the "world's smallest turbine" which Leonid Sazanov calls "one of the most important enzymes on earth." His team's work focuses on the rotary portion of the enzyme (Fo), which is hidden inside the lipid bilayer.Our structure of bovine dimers has a wedge made of small proteins and specific lipids in the membrane domain of each monomer that imposes a range of acute angles on the central axes of the monomers, and a pivot between the wedges accommodates rocking motions of the machine accompanying catalysis and other movements that happen independently. It also throws light on how the membrane rotor is made to turn.
They also figured out a special plug that can halt the machine when the cell is dying. Called the "permeability transition pore," this plug can only be opened by a "hook apparatus" that is connected to the catalytic portion in F1 and to the bottom of the membrane. Calcium signals trigger a large conformational change, which yanks on the hook and opens the plug. Protons leak out, destabilizing the rotor, and halting the machine. It sounds like an emergency stop switch.To solve the structure of the Fo domain and the entire complex, the researchers studied the enzyme from sheep mitochondria using cryo-electron microscopy. And here, ATP synthase poses a special problem: because it rotates, ATP synthase can stop in three main positions, as well as in substates. "It is very difficult to distinguish between these positions, attributing a structure to each position ATP synthase can take. But we managed to solve this computationally to build the first complete structure of the enzyme", Sazanov adds.
Bacterial Flagellum
More has been discovered as well about a familiar ID standby, the bacterial flagellum. European scientists led by Vitan Blagotinsek, also publishing in PNAS,2 found that "An ATP-dependent partner switch links flagellar C-ring assembly with gene expression." This relates to controlling how many flagella the cell constructs on the surface. Some bacteria have one; others have two or more. How the cell regulates the number of flagella has been "only poorly understood" before now.
In the film Unlocking the Mystery of Life, Scott Minnich referred to feedback mechanisms that control how many parts are sent to the construction site. The European team elucidated one such feedback. The secret to controlling how many flagella are constructed, they found, occurs when an ATP-powered part named FlhG flips a molecular switch between two other components:
Depending on its ATP-dependent oligomerization state, FlhG interacts either with the C-ring protein FliM during flagellar assembly or with flagellar master regulator FlrA. This partner switch between FliM and FlrA establishes a regulatory network critical for the numerical regulation of flagella, in which the physical assembly of the flagellum transcriptionally feeds back to prevent the production of more building blocks.
Comment: From the above mentioned film Unlocking the Mystery of Life :
New information was also obtained about the pentagon-shaped cap that controls orderly addition of monomers to the growing propeller (see the animation "Darwinian Evolution vs Bacterial Flagella Motor and Hook Mechanism" here at 1:00 through 2:15). Publishing in Nature Communications,3 Al-Otaibi and six others in the UK used cryo-electron microscopy to study the "structure of the bacterial flagellum cap complex" that "suggests a molecular mechanism for filament elongation."
Comment: Darwinian Evolution vs Bacterial Flagella Motor and Hook Mechanism:
The pentagonal cap, looking somewhat like a stool, is composed of FliD proteins. This cap is first extruded in pieces through the central channel of the hook. Its subunits self-assemble at the end, while forming "an extensive set of contacts across several subunits, that contribute to FliD oligomerization." The cap then guides individual flagellin proteins coming up through the channel into their positions in the growing filament, as shown in the animation. This is a well-controlled process, they found, dependent on the precise arrangement of amino acids in each protein, so that contacts work as intended.
As for the specificity of this operation, the scientists say that "disrupting these interactions has a significant effect on cell motility." One recalls the words of Michael Behe, "If any of these parts are missing, then either you get a flagellum that doesn't work, because it's missing the hook or it's missing the drive shaft or whatever, or it doesn't even get built within the cell."Once outside of FliD, we propose that exposed hydrophobic residues act as a chaperone, and promote flagellin folding in its insertion site. In order to accommodate the next flagellin subunit, conformational changes need to occur to open an adjacent binding pocket. We propose that the folding of the new flagellin protomer leads to dislodging of the cap complex, that rotates by ~65°, thus positioning an adjacent cavity of the cap complex close to the next flagellin insertion site (Fig. 5). This hypothesis agrees with previously proposed mechanisms of flagellar elongation.
ID advocates should feel encouraged to see arguments for irreducible complexity standing the test of time. After more than two decades, evolutionists still "have failed to offer any detailed Darwinian explanation" for a rotary motor arising by natural selection, as evidenced by the complete silence of all three papers on that key question.
Notes
- Spikes, Montgomery and Walker, "Structure of the dimeric ATP synthase from bovine mitochondria." PNAS September 8, 2020. https://doi.org/10.1073/pnas.2013998117.
- Blagotinsek et al., "An ATP-dependent partner switch links flagellar C-ring assembly with gene expression." PNAS August 25, 2020 117 (34) 20826-20835. https://doi.org/10.1073/pnas.2006470117.
- Al-Otaibi et al., "The cryo-EM structure of the bacterial flagellum cap complex suggests a molecular mechanism for filament elongation." Nature Communications June 25, 2020: 11, 3210 (2020). https://doi.org/10.1038/s41467-020-16981-4.



Comment: The following animations illustrate the incredible complexity and functionality of the molecular machines that reside in humans: