Junk DNA
ID scientists have been disputing "junk DNA" claims for many years now. Another supportive case came to light recently. The Simons Foundation found that mutations in non-coding DNA (once considered evolutionary leftovers, or junk) can be implicated in the development of autism. While a diagnosis of autism can be devastating for a family, the important point is that transcripts of non-coding DNA, even if they do not yield proteins, still influence the health of the individual. The Simons Foundation calls this discovery a first.
Research into the genes of 1,790 patients not having a prior family history of autism led to the inference that the condition was due to mutations. Notice how the researchers say that "junk DNA" is a misnomer, and that the implications of their finding extend beyond this particular condition.Leveraging artificial intelligence techniques, researchers have demonstrated that mutations in so-called 'junk' DNA can cause autism. The study, published May 27 in Nature Genetics, is the first to functionally link such mutations to the neurodevelopmental condition. [Emphasis added.]
By "non-inherited mutations," the scientists refer to spontaneous mutations in that individual, not mutations passed down from the parents. What the research shows is that mutations in so-called "junk" DNA can also be implicated in diseases as diverse as cancer and heart disease. The non-coding DNA, therefore, is not "genetic dead weight," but has functional importance even if it is not transcribed into proteins. The lead scientist essentially calls the "junk DNA" myth a science stopper:The analysis predicted the ramifications of genetic mutations in parts of the genome that do not encode proteins, regions often mischaracterized as 'junk' DNA. The number of autism cases linked to the noncoding mutations was comparable to the number of cases linked to protein-coding mutations that disable gene function.
The implications of the work extend beyond autism, Troyanskaya says. "This is the first clear demonstration of non-inherited, noncoding mutations causing any complex human disease or disorder."
Irreducible ComplexityTroyanskaya says she and her colleagues will continue improving and expanding their method. Ultimately, she hopes the work will improve how genetic data are used for diagnosing and treating diseases and disorders. "Right now, 98 percent of the genome [i.e., the non-coding DNA] is usually being thrown away," she says. "Our work allows you to think about what we can do with the 98 percent."
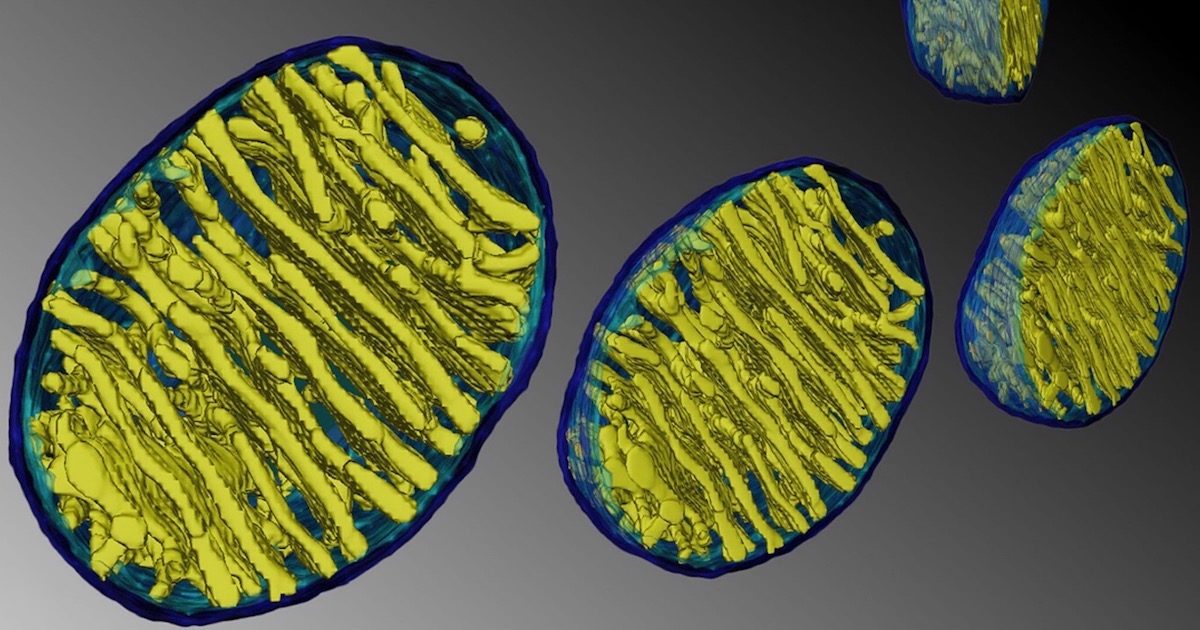
Meet TOM. He's an important guy. He stands for "translocons of the outer membrane" - a group of transporters who move proteins to another location through the outer membrane of mitochondria. The powerhouses of cells, the mitochondria, protect their valuable energy motors like ATP synthase behind two membranes, inner and outer. TOM has a partner named TIM who translocates proteins through the inner membrane. Like border guards, TIM and TOM escort proteins through these checkpoints, authenticating each passenger using established protocols. As a paper in Nature explains, sometimes the border guards get stressed by overcrowding.
Mitochondria contain a minimal genome that builds some of the proteins they need, but many other proteins are encoded in the nucleus and translated by ribosomes in the cytoplasm. Those precursor proteins must pass through TOM's entry gates before folding inside the mitochondrion. Sometimes, however, they get stuck in the gates, forming dangerous gridlocks downstream with disastrous consequences. Fortunately, another protein knows what to do. Called Ubx2, this "quality-control" protein knows how to unclog the gate when one individual gets "arrested" and stopped in the passageway. But Ubx2 doesn't work in isolation:Mitochondrial biogenesis and function depend on the import of more than 1,000 proteins that are synthesized as precursors on cytosolic ribosomes. The TOM complex forms the entry gate for almost all mitochondrial precursor proteins. Impaired mitochondrial protein import triggers a number of cellular stress responses that are coordinated by a transcriptional regulatory pathway (known as mitoprotein-induced stress response).
As with humans, there are many guys named TOM. The TOM family includes Tom20, Tom22, Tom40, Tom70 and others, each with specific roles. The authors of the paper use the phrase quality control 10 times, and evolution zero times - not surprising, since the translocation process is irreducibly complex. Numerous components work together on an essential process in the cell - power generation - and without all of the components functioning properly, the cell would die of stress.A pool of Ubx2 binds to the TOM complex to recruit the AAA ATPase Cdc48 for removal of arrested precursor proteins from the TOM channel. This mitochondrial protein translocation-associated degradation (mitoTAD) pathway continuously monitors the TOM complex under non-stress conditions to prevent clogging of the TOM channel with precursor proteins. The mitoTAD pathway ensures that mitochondria maintain their full protein-import capacity, and protects cells against proteotoxic stress induced by impaired transport of proteins into mitochondria.
Human Uniqueness
According to the standard evolutionary account, humans and chimpanzees evolved from a common ancestor. But when the genomes are compared, even if one were to accept the incorrect value of 2 percent difference, that difference amounts to millions of base pairs that had to mutate and be selected. Now, Science Daily reports on findings at the University of Toronto that show even more differences. "Dozens of genes previously thought to have similar roles across species are in fact unique to humans," the subtitle reads. Those newly identified genes are called transcription factors (TF). Transcription factors were thought to be largely unchanged between humans and fruit flies.
The phrase "rapidly evolving" can be understood as theory-laden jargon representing Darwinian assumptions. The researchers publishing in Nature Genetics accept those assumptions, but have to conclude that organisms have more disparate TFs than previously thought. Using a mathematical technique called similarity regression, they warn their colleagues that "the assumptions, they are a-changin'."As for TFs that have unique human roles, these belong to the rapidly evolving class of so-called C2H2 zinc finger TFs, named for zinc ion-containing finger-like protrusions, with which they bind the DNA.
Their role remains an open question but it is known that organisms with more diverse TFs also have more cell types, which can come together in novel ways to build more complicated bodies.
All of us can breathe a sigh of relief that we are not as similar to fruit flies as evolutionists had led us to believe. We have unique genomes, we have TOM helping operate our quality control, and the junk they told us we were hoarding has a purpose after all.Similarity regression inherently quantifies TF motif evolution, and shows that previous claims of near-complete conservation of motifs between human and Drosophila [fruit fly] are inflated, with nearly half of the motifs in each species absent from the other, largely due to extensive divergence in C2H2 zinc finger proteins. We conclude that diversification in DNA-binding motifs is pervasive, and present a new tool and updated resource to study TF diversity and gene regulation across eukaryotes.



"Junk DNA", hey, hmm, I thought we'd accounted for all DNA, to date.
Environment rules, unless something else?Wishes to murky the waters of the DNA pond.
"GM modified", what does that suggest?
Messing with Nature, that's what it means.
"GM" should be a crime, yes, one against humanity.
If, and it's a big "if", you support the concept of "intelligent design", then if it's not of God's making, explain the complex interface, that is that governing intelligence, that so communicates, and assembles order, out of chaos.