Murphy is a "hunter-gatherer" - a biologist charged with cataloguing Earth's rich array of plants and animals. For decades, he has plunged into the farthest-flung corners of the globe to find and collect new species. "It's not for everyone," he says. "People can end up with broken bones or malaria or puff up with insect bites, and the days are long and tough." Indeed, the dangers can be life threatening. In 2001, Murphy's friend and fellow collector Joe Slowinski died after being bitten by a venomous snake he had caught in Myanmar.
Despite the risks, hunter-gatherers will soon be in high demand as an audacious scheme gets under way. This biological "moonshot", known as the Earth BioGenome Project, is scheduled to launch in June. Its mission is to sequence the genomes of all known species of flora and fauna on Earth. Nature's recipe books could hold clues to making far superior medicines, materials, biofuels and crops, unravelling our evolutionary past and help us to be better custodians of our planet. The first challenge, however, will be collecting specimens from the wild. Then comes the sequencing itself, which will require Herculean amounts of human labour and computing power. Can it be done?
The Human Genome Project seemed equally far-fetched when it was proposed in the late 1980s. "There were many people who told us, 'This is a waste of money, it's way too costly'," says David Haussler at the University of California, Santa Cruz. It cost $2.7 billion - or about $4.8 billion at today's prices - and took over a decade to complete, but the treasure trove of information it unlocked has wildly exceeded expectations. Not only did it give birth to the personalised medicine revolution, it also propelled advances in diverse fields including forensics, archaeology and bioinformatics. Not to mention, every $1 of public money invested has since generated $141 in economic activity. "It's paid for itself many times over," says Haussler.
It was this success that inspired biologist Harris Lewin at the University of California, Davis, to start pondering an Earth-scale genome project three years ago. "Everybody's first expression is like, 'He's gone insane'," he chuckles. But his initial rough calculations suggested it was doable. "I saw that with today's technology, the time and cost would basically be the same as for the Human Genome Project," he says. "The insights we've gained from just one genome have been incredible, so imagine what could be revealed by sequencing the rest of life?"
At the time, Lewin was a member of Genome 10K - a scheme launched in 2009 with the goal of sequencing 10,000 vertebrate genomes. Similar projects soon popped up aiming to sequence 10,000 bird genomes (B10K), 5000 insect genomes (i5K), 10,000 dog genomes (Dog 10K), 7000 marine invertebrate genomes (GIGA) and 1000 plant genomes (1KP). "It just seemed like the logical next step to sequence everything," he says.
In November 2015, at a meeting of 23 biologists at the Smithsonian Institution in Washington DC, Lewin floated his idea: to sequence, over a 10-year period, every eukaryote known to exist on Earth. These are organisms with cell nuclei, including animals, plants and fungi - of which there are about 1.5 million. "There were some very sober people there, but by the end of the meeting, enough were convinced," he says. A framework for the Earth BioGenome project was teased out over several follow-up meetings, and partnerships were forged with research powerhouses including the Smithsonian Institution, the Wellcome Sanger Institute in the UK, BGI in China, and the São Paulo Research Foundation (FAPESP) in Brazil.
Just 2500 eukaryotes have been sequenced to date, so it will be a gargantuan endeavour. The goal for the first three years is to produce a high-quality genome for one member of each of the 9000 eukaryotic families. Then, over the following three years, a draft genome for one member of each of the 150,000 genera - the taxonomic group below families - will be assembled. And the last four years will be spent compiling drafts for the remaining species, which can be refined later on.
These lofty targets mean sequencing eight high-quality genomes per day during the first phase, more than 100 drafts daily in the second, and in excess of 1000 every day in the third. With current technology, a high-quality genome takes about a week to sequence and costs between $1000 and $30,000, depending on its size. A draft takes a few hours and cost about $800. But costs and sequencing times are likely to fall as technology improves, says Lewin. And, despite the enormity of the task, Guojie Zhang, a biologist at BGI, thinks it can be done. "With the sequencing power we can access now, it would actually be possible to finish the sequencing for all 1.5 million eukaryotic species within a year," he says.
In fact, the main difficulties will be in logistics, including getting permission from governments to sequence native species, and collecting and preparing samples. Some specimens will come from museum collections, but they are only suitable for draft genomes because high-quality sequences require fresh tissue from multiple organs - hence the need for hunter-gatherers like Murphy, who is based at the University of Toronto in Canada and collects specimens for Genome 10K. Finding rare species - particularly in difficult-to-access areas like the deep sea or dense forest - will be challenging. Murphy's speciality is frogs and snakes. Sometimes he travels by plane or boat to reach remote places, but mostly he treks on foot into the wilderness with local porters or elephants carrying his supplies. The investment of time and effort can be huge. "We expect one of the legless lizards we want for Genome 10K may take six months of fieldwork to find, if we're lucky," he says.
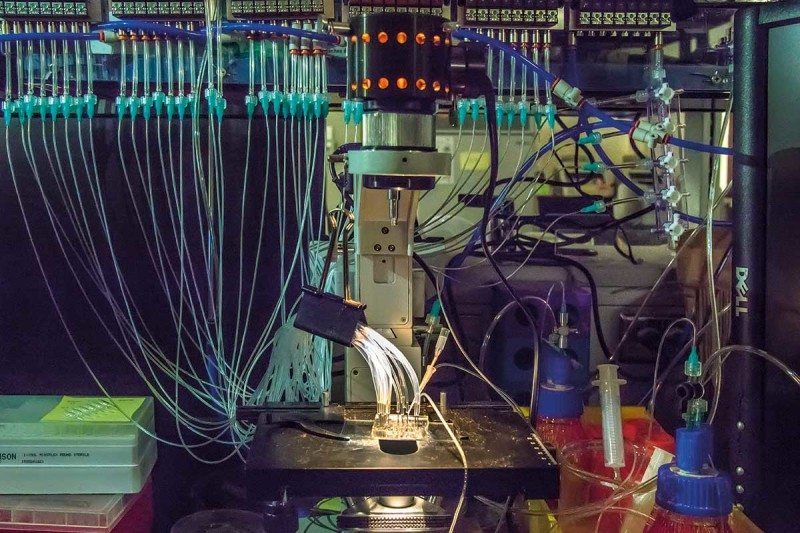
Another challenge is how to store the whopping amounts of data. It is estimated that the project will generate several thousand petabytes per year - more than all the videos uploaded to YouTube annually. Again, this could be a spur for innovation, this time in bioinformatics. One option may be to store DNA data in... DNA. The code's letters A, T, C and G can be used like the 0s and 1s in regular computing, and researchers at Columbia University recently showed that a gram of DNA can encode 215 petabytes of digital data.
Finally, the project's founders want to make sure it benefits all involved fairly. To do this, they have signed up Peruvian entrepreneur Juan Carlos Castilla-Rubio to build the Earth Bank of Codes. This open-access database will record the genomic sequence, appearance, location and associated indigenous knowledge for each species. The data will go on a blockchain - a type of ledger used in cryptocurrency - that traces where and how the information is used. Any commercial benefits can then be shared appropriately with all contributors, including local people who provide traditional know-how.
Castilla-Rubio came up with the idea when he was looking for ways to shift the Amazon's economy away from destructive industries like farming, logging and mining towards more knowledge-based enterprises that preserve the environment. He recognised the huge economic potential of the genomic information tied up in the Amazon - which is home to 15 per cent of Earth's land biodiversity - but also the problem of exploitation. In the past, large corporations have tapped the region's natural resources and indigenous knowledge without paying their dues, for example, when a blood-pressure medication was developed from snake venom traditionally used by Amazon people on their arrow tips. The blockchain approach should prevent this type of biopiracy and ensure that the Earth BioGenome Project sticks to the Nagoya Protocol - an international agreement made in 2010 to recognise the rights of countries over their genetic resources and traditional knowledge.
Of course, all this must be paid for - the total cost is estimated at $4.7 billion. As yet, the project has no dedicated funding, although publicity at this year's World Economic Forum in Davos, Switzerland, has sparked enthusiasm. "Many individual countries are now expressing interest or are close to making large commitments, and we're talking to some prominent people," says Lewin. But existing sequencing drives such as Genome 10K do have funding, and when the project is officially launched in June, it will begin by building on these. Lewin and fellow project leaders Gene Robinson at the University of Illinois and John Kress at the Smithsonian Institution plan to start by coordinating the activities of the various schemes to make sure enough genomes are sequenced each year to meet overall targets.
Given all the expense and effort, what pay-off can we expect? Lewin is confident that the open nature of the Earth Bank of Codes will lead to discoveries and innovations all around the world. One area with huge potential is pharmaceuticals. Already, about half the world's drugs are natural products or derivatives - including aspirin and Botox - and we have only just scratched the surface. Genome sequencing can inspire new medicines by revealing how plants and animals have evolved their sophisticated defences against predators and disease. Guilherme Oliveira at the Vale Institute of Technology in Brazil, for example, is sequencing the Amazon's jaborandi tree, which produces pilocarpine, a drug used to treat the eye disease glaucoma. Once his team has done this, they will be able to work out the pathway that produces the valuable chemical. It may be possible to replicate this process synthetically or tweak it to make even more potent medicines.
Another major beneficiary will be conservation, says Haussler. He believes sequencing endangered species will give us clues about which are most vulnerable to climate change and need the most attention. This knowledge will help caretakers manage remaining populations too. For example, when researchers sequenced the genome of the critically endangered Californian condor, they found a recessive gene that was causing fatal skeletal abnormalities in some of the 400 remaining birds. Breeders are now using this information to selectively match up individuals without this gene to improve the health of the population.
Sequencing all life will also let us retrace evolution and see where each species sits in the family tree, says Susan Brown, a biologist at Kansas State University. This will answer long-standing questions such as whether vocal learning evolved once or multiple times in birds. Already, DNA sequencing has revealed unexpected relationships in our family tree. For instance, we have discovered that the same genes regulate circadian rhythms in both plants and animals. "In the same way that the periodic table shows you how the different elements are related, the tree of life reveals relationships between different species," says Brown.
Lewin believes the potential benefits of the project go far beyond biology. The wealth of genomic information is likely to find applications in all sorts of fields from renewable energy and engineering to agriculture and artificial intelligence, he says. There could also be benefits we can't even conceive of yet. In the same way we couldn't predict all the innovations that came out of the Human Genome Project, "we don't know what we don't know", he says.
Such enthusiasm is what keeps Lewin criss-crossing the globe to promote the Earth BioGenome Project. He is well aware there are vast challenges ahead, but he is also certain they will be worth it. "Sometimes, you just have to go for these things," he says. "We've got the technology, we've got the expertise, now we just need the will."
Megaprojects for microorganisms
If you thought 1.5 million eukaryotes was a lot of genomes to sequence, the number of prokaryotes will blow your mind. It is estimated that there are up to 1 trillion species of these microorganisms, which include bacteria and archaea, and we have only classified a few thousand so far. The reason for this slow progress is that prokaryotes are hard to isolate. Most can survive only in the precise conditions of their natural habitat - be it a hydrothermal vent or a cow's gut - so cannot be grown and studied in the lab.
The game changer is metagenomics. This technique allows us to sequence all the DNA in a sample taken from an environment such as seawater, soil or faeces, and then pull it apart to identify the individual species. The biggest metagenomics project yet is currently under way at the US Department of Energy's Joint Genome Institute. It is set to publish the genomes of more than 100,000 species of bacteria and archaea from a range of different environments this year.
The information contained in prokaryotic genomes could help us develop novel antibiotics, because bacterial DNA contains blueprints for chemicals to fight off other bacteria. It may also contain instructions on how to break down pollution, produce industrial chemicals, improve food production and much more.




Technology is all good, technology is the miracle.
No one has ever been hurt by too much technology, that is absurd.
A society pushing for more technology is just like a man (or woman) pushing for more and more money.
No one has ever been hurt by having too much money, that is absurd.
When you have a lot of money, and it is yours, you have to guard it, so no one else takes it. And this is good, it keeps you busy and growing, plotting and scheming.
So when you guard the money you have, which is a lot, you need to get more money, which is a more lot. To guard the money that you have, which was a lot, but now is not enough. So you get more, to guard and protect the lot that you have.
This is all good and great for the economy. Keeping things busy and growing, plotting and scheming.
Technology is all good, technology is the miracle.
And it will create endless miracles, and be great for the economy.
All you have to do, is guard it successfully from thieves and poachers and hackers and the like.
You will live in Lalaland forever, which is good, all good.
I repeat: No one has ever been hurt by too much technology.
Technology is all good.
And you cannot and will not be hurt by having too much of it. Just be sure to hang on to every last bit of it and always be conspiring to get more.
Lalaland forever!
DUH.
FUCKING FUCKING FUCKING DUH.
ned,
OUT