But when you examine the paper supposedly supporting Behe's claim, you find, argue Lents and Hunt, that about half of them don't seem to have any damaging mutations, and that perhaps "none of the 17 most positively selected genes in polar bears are 'damaged'."After replying to two objections from Lents and Hunt, we now know it's quite a stretch for Coyne, Lents, or Hunt to suggest that "none" of the polar bear genes may have been damaged, especially when Behe's authority, Liu et al. (2014), concluded that "a large proportion (ca. 50%) of mutations were predicted to be functionally damaging," and given that Lents and Hunt themselves admitted at least seven genes were "unequivocally predicted to possess at least one 'damaging' mutation."
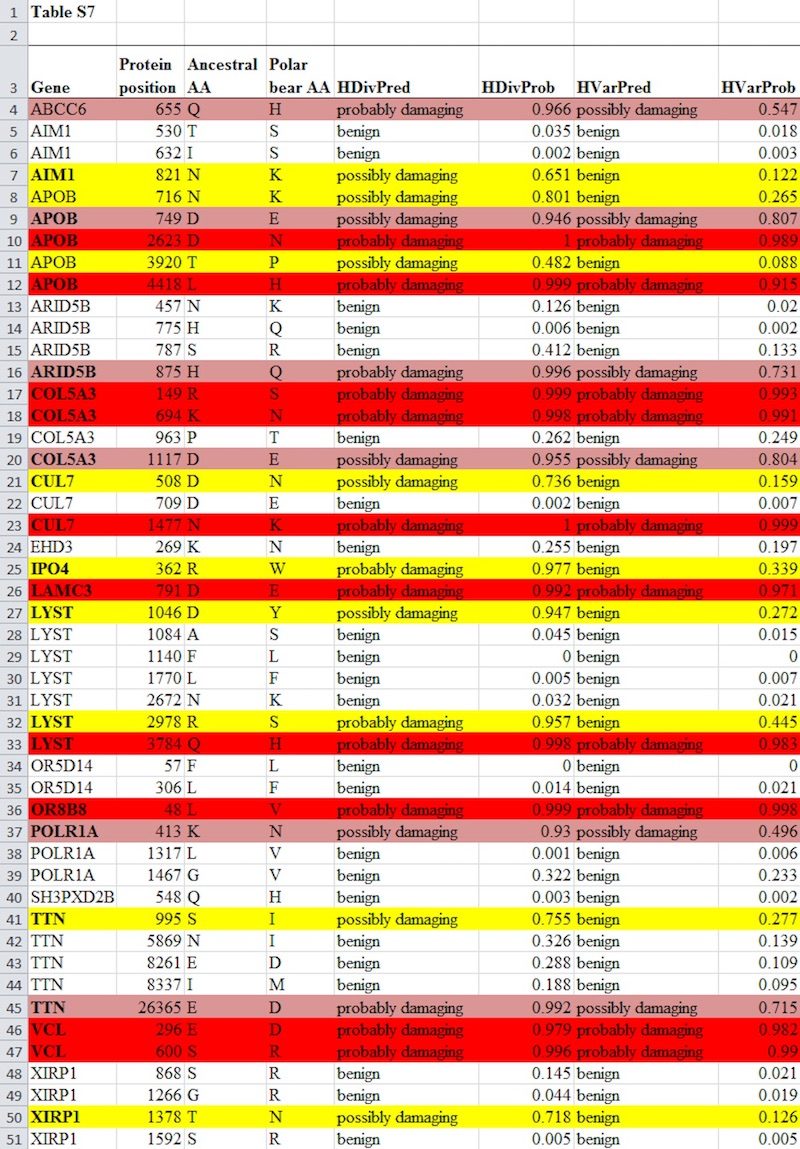
Behe also wrote his own rebuttal to Coyne, Lents, and Hunt where he provided the data that he relied upon in Darwin Devolves to conclude that 14 out of the 17 genes had experienced mutations that were probably damaging. Behe listed all of the mutations in Table S7 of Liu et al. (2014) that were predicted to be damaging, as well as the genes they occurred in, and stated, "Below is the relevant information from Liu et al.'s Table S7. Those who can understand the table will see that it supports every actual, undistorted claim I made about the polar bear." However, Behe did not print Table S7 in its entirety; he only printed the data needed to respond to the question at hand and show those 14 genes experienced degradative mutations. He did, though, link to the full article so people could view the entire table.
An Uproar from the Critics
This led to an uproar from Behe's critics. Lents charged Behe with being "deeply misleading" and "intentionally leaving out evidence that is contrary to his position," although Lents acknowledged that Behe "isn't lying exactly" because, Lents conceded, Behe said he was just showing the "relevant information." Here's what Lents said:
In Behe's defense, he doesn't explicitly say that he's presenting the whole Table. So he isn't lying exactly. Instead, he says that he is presenting "the relevant information" from the Table. I find this deeply misleading. This whole discussion is about the nature of adaptive mutations in the evolution of species and Behe's arguments is that most of them are damaging. By presenting only the mutations that are predicted to fit that argument, he is intentionally leaving out evidence that is contrary to his position.Lents claimed that Behe's "selective presentation of the chart leaves out all of the alterations that are not predicted to damage the protein in which they occur," charging that "he slices it up with surgical precision so that he can present 'the relevant information,' that is, the information that appears to support his position." Elsewhere Lents charged that Behe "doctored the hell out of a chart."
Apologies for the Repetition
We've already shown the full, unedited Table S7 twice in this series, and we'll show it once more here just below. Sorry for the repetition. But before explaining why the data that Behe didn't list in no way challenges his position, it's important to remember what was being debated here. It easily explains why Behe showed the parts of the table that he did:
- In Darwin Devolves, Behe claimed that the data from Liu et al. (2014) show that as many as 14 out of 17 strongly selected genes in polar bears probably experienced at least one damaging mutation.
- Nathan Lents, Arthur Hunt, and Jerry Coyne disagreed with Behe, and challenged his claim that so many polar bear genes experienced damaging mutations. These critics said the actual number of genes with degradative mutations might be as low as seven, or even zero.
- Behe replied to these critics by listing all of the mutations that were predicted to be "damaging" from Liu et al.'s Table S7. If you count up the number of genes affected by the mutations Behe listed, it's 14 genes, just as Behe wrote in his book. Behe provided exactly the data needed to rebut the challenges from Lents, Hunt, and Coyne. In his own words, he provided the "relevant information" to answer their challenge.
Now Lents claims that Behe is "intentionally leaving out evidence that is contrary to his position." And what exactly is Behe's position? It is that adaptive mutations are usually damaging, and are only rarely "constructive." His thesis also explicitly grants that neutral or benign mutations that are non-adaptive are common. Did the data from Table S7 that Behe didn't list contradict his thesis? Not in any way.
First, much of the data from Table S7 that Behe didn't list entails results from algorithms that indicated mutations were damaging! Specifically, Behe didn't list the results of 16 analyses of PolyPhen-2 that predicted mutations were damaging - evidence that presumably supports his thesis. For what nefarious reason did Behe leave this data out? None, of course. He didn't list this data because the chart is quite long and the data that he did list was entirely sufficient to validate his arguments and rebut his critics. Recall that Liu et al. found that if just one algorithm predicted that a given mutation was damaging, then that qualified the mutation as probably being damaging. Behe too felt that if one algorithm found a mutation was damaging, there was no reason to list the results of multiple analyses.
Second, Behe's critics are making a big deal about his not showing the data in the table where mutations were predicted to be "benign" - but benign or neutral mutations don't challenge his thesis. Data only challenges Behe's thesis when a mutation is shown to be constructive. In this regard, it's crucial to point out that the mutations that Behe didn't list from the chart that were not said to be damaging were also NOT said to be "constructive." They were said to be "benign." That's a key point that completely undermines Lents's charge that the data Behe doesn't list is somehow "contrary" to his position.
As Lenski Admits
Saying that the mutations are "benign" is not equivalent to saying that they are "constructive" or somehow improved function. From Liu et al., we have evidence that many mutations are probably damaging or probably benign, but that paper does not claim to provide any direct evidence that the mutations labeled "benign" are constructive. Indeed, Richard Lenski admitted that "The program [used by Liu et al.] simply cannot detect or suggest that a protein might have some improved activity or altered function." Thus it's impossible that any of the data in the table, listed or unlisted, could show Behe hiding evidence of constructive mutations, because the program does not report constructive mutations. Rather, the benign mutations that Behe didn't list are arguably data points that don't really say anything for or against Behe's thesis.
Is it a problem for Behe's thesis if not all the mutations were degradative? Not at all, for Behe happily recognizes that neutral mutations not only occur, but are common. As Behe writes:
What studying neutral evolution can't do is tell us what caused an organism to adapt - the most critical question in this book since, by definition, neutral mutations have no effect on a species's survival. In fact, since neutral mutations are the bulk of changes at the molecular level, they substantially obscure evolutionary changes that do affect species. (Darwin Devolves, p. 98, emphasis added.)Behe allows that neutral mutations are very common - "the bulk of changes at the molecular level" - yet from the data reported in Table S7 of Liu et al. (2014), it's quite probable that the "benign" mutations are simply neutral, meaning not constructive. Indeed, one paper on PolyPhen-2 explains that when the program labels a mutation as "benign" that means "most likely lacking any phenotypic effect" - i.e., neutral. So the data that Behe doesn't list isn't the kind that would challenge his thesis - i.e., constructive mutations. Rather it shows mutations that aren't damaging and most likely had a neutral effect.
Another Behe Critic
The reason Behe didn't show the entire chart is the same reason that another Behe critic, Joshua Swamidass, didn't show the entire chart on a recent webinar: saving space. In the webinar, Swamidass repeated the arguments of Lents and Hunt and attacked Behe for not showing the entire table, arguing Behe "did not correctly represent the conclusions of the scientists," which shows that he "misrepresents science and ignores evidence against him." Swamidass concludes, "Over and over again I don't really know if I can trust where he's coming from."
Comment: Swamidass and the Darwinists like him are either confused or malevolent. It's not a good sign that they are seemingly hallucinating controversy and "misrepresentation" where there is none. But even if it is just hallucination prompted by cognitive dissonance - and not a cynical attempt to maliciously smear Behe - they could have merely noted their own confusion by asking if there was an alternative explanation for what they thought they were seeing. Instead, they impute mendacious motives to Behe. Perhaps they really are projecting.
The full chart is some 49 lines long and 8 columns wide in Excel, and it's far simpler to show a representative part of a huge chart that proves your point than to show the entire thing. It's reasonable that Swamidass only shows the first 29 lines of the chart, leaving off about 40 percent of the chart. For the point he was trying to make, it was enough to just show the first 60 percent. Much like Behe, Swamidass indicated that he wasn't showing the entire chart.
Perhaps it would have been better for Behe to have shown the full Table S7 if for no other reason than to avoid the potential for contrived accusations. In any case, for anyone who still thinks that there's something to hide, here's Table S7 in full, for the third time:
The data from this table that Behe listed in his response here comprise those instances when the HDiv algorithm predicted that a mutation was possibly or probably damaging (this is seen in the HDivPred column). He did not list the results of the HVar algorithm nor mutations where HDiv did not predict a mutation was damaging (i.e., benign). This was reasonable, because the data Behe listed allowed the reader to validate his most aggressive estimate in Darwin Devolves of the proportion of positively selected polar bear genes that probably experienced at least one damaging mutation. That number was 83 percent, and it can be reproduced by counting the number of genes in the table above which have a mutation that is predicted by HDiv to be probably or possibly damaging. This comes to 14 genes, which is 83 percent of the 17 total genes investigated. As was already discussed here and here, Behe's counting method for his upper estimate followed the precise protocol used by Liu et al. (2014). Thus, the data from Table S7 that Behe listed in his post was sufficient to validate his highest estimate of the number of polar bear genes consistent with his "devolution" hypothesis.
We've already explained why the "benign" mutations that weren't predicted to be damaging in Table S7 that Behe didn't list did not contradict his thesis. Maybe it's unfortunate that Behe did not list the results of the HVar algorithm from Table S7 - again, not because he was hiding contrary data but because listing the HVar results would have allowed the reader to also validate how he arrived at his minimum estimate in Darwin Devolves of the number of polar bear genes that experienced degradative mutations (65 percent).
The preceding post in this series explained why Behe's minimum estimate was accurate. One can confirm this by counting mutations in the table above where both the HVar and HDiv algorithms found the mutation was possibly or probably damaging. This shows 11 genes experienced such a "damaging" mutation, which corresponds to Behe's lower estimate that at least 65 percent (11 out of 17) of positively selected polar bear genes experienced a damaging mutation. You can't validate his lower estimate unless you see the results of both HVar and HDiv, but the point is this: The HVar results from Table S7 that Behe left out of his post were not "contrary to his position" but rather validate his minimum estimate of damaged polar bear genes in Darwin Devolves.
Contradictory Criticisms
So Behe is being subjected to self-contradicting criticisms:
- On the one hand, as seen here, the critics say Behe can't use the data in Table S7 to support his thesis because the program can't rule out the possibility that the mutations being studied are constructive.
- On the other hand, they say Behe is hiding data in Table S7 that contradicts his thesis.
The first option is wrong because the PolyPhen-2 program does not give absolute answers but probabilistic ones. Even if there's a small possibility that some mutations may have been constructive but were wrongly tagged as damaging, the best evidence suggests the mutations were probably degradative. This means that Behe is justified in citing the evidence from Liu et al.'s PolyPhen-2 analysis reported in Table S7. It shows that the best evidence indicates many mutations in polar bear genes were probably damaging to the gene's function. See the second installment in this series.
The second option is wrong because the data that Behe is accused of hiding (the benign mutations and the results of the HVar algorithm) are fully consistent with his thesis, and even support his claims:
- Regarding the "benign" mutations that Behe didn't list, the PolyPhen-2 program gives no reason to think that they are constructive, and every reason to think that they are neutral mutations. Behe fully allows that non-adaptive neutral mutations are extremely common. It's not the non-adaptive neutral mutations that concern Behe's thesis, but the adaptive ones. And as Darwin Devolves shows, adaptive mutations tend to be degradative rather than constructive.
- Regarding the results of the HVar algorithm, this data also does not contradict Behe, but in fact validate his minimum estimate of the proportion of positively selected polar bear genes that probably experienced a damaging mutation.
Comment: Maybe it's time Lents grew up.




Comment: