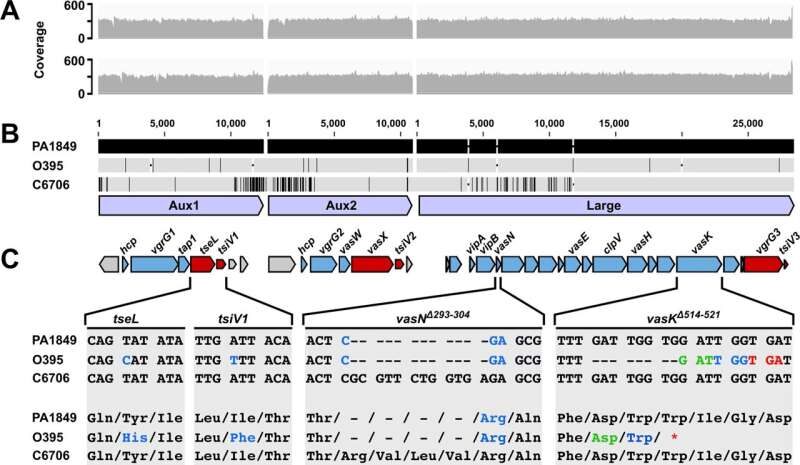
© DOI: 10.1038/s41467-021-26847-yFig. 1: The T6SS gene clusters of the classical V. cholerae strain PA1849. A Illumina-generated reads were mapped to a 6th pandemic classical strain (O395) and a 7th pandemic El Tor V. cholerae strain (C6706). Read coverage plotted against the nucleotide sequence of the three T6SS gene clusters (shown as light blue arrows in panel (B)) separated by white vertical lines that indicate the intervening genomic sequences not included in this analysis. The top plot (gray) represents read coverage against the O395 reference, and the bottom plot (gray) represents read coverage against the C6706 reference. B Nucleotide alignment of the three T6SS clusters (bottom light blue arrows) from PA1849, O395 and C6706. The top black bars, representing the PA1849 T6SS clusters, are designated as the reference sequences. Conserved residues in O395 and C6706 sequences are represented by gray bars with single-nucleotide polymorphisms (SNPs) highlighted by vertical black lines and insertions or deletions (INDELS) of base pairs represented by gaps with horizontal black dashes. C A schematic gene map for each T6SS gene cluster displays structural genes as blue arrows and effector/immunity pairs as red arrows. Zoomed alignments of regions of interest in strains C6706, O395 and PA1849 are shown. Hyphens in the alignment indicate deleted nucleotides (above) and amino acids (below). Nucleotides and their corresponding amino acid changes are colored. Multiple colors are used to indicate the new reading frame for the vasK frameshift mutation.
How did the modified cholera strains develop and spread, and what might have contributed to their success? Scientists from the Max Planck Institute for Evolutionary Biology in Plön, Germany, and CAU Kiel, in an international team with colleagues from City College New York and the University of Texas Rio Grande Valley, have now gained new insights into a molecular mechanism that provides insight into the interactions between cholera bacteria and may have played a role in the emergence of the seventh pandemic.
In their natural environment, bacteria are subject to competition with other bacteria for space and nutrients. In this process, molecular mechanisms help them to hold their own. One such mechanism is the so-called "type 6 secretion system" (T6SS), with which a bacterium transports toxic proteins into a neighboring bacterium and thereby kills it. Thus,
cholera bacteria of the seventh pandemic use their T6SS to keep other bacteria in check and presumably more easily cause infection.
Researchers now had the special opportunity to study the T6SS of cholera bacteria from previous pandemics. For this purpose, among other things, the T6SS genome sequence of cholera bacteria from the 2nd pandemic was reconstructed from a museum specimen from the 19th century in a complex procedure and recreated in the laboratory.
In the process, the scientists
were able to show that 2nd and 6th pandemic cholera bacteria lack a functional T6SS. As a result, the bacteria of earlier pandemics not only lack the ability to attack other bacteria, they are themselves killed by bacterial strains of the seventh pandemic. This may have been one of the reasons that older cholera strains were displaced by modified cholera strains of the seventh pandemic and are now hard to find.
Data from new labDaniel Unterweger, one of the study's authors and a group leader at the Max Planck Institute in Plön, Germany, says: "With these findings, we
support the theory that microbial competition between bacteria is very important for understanding pathogens and bacterial pandemics. Our research on the cholera bacterium was made possible by an S2 laboratory newly established at the institute. Here, we can conduct experiments with bacterial pathogens under the necessary safety precautions. The study contains some of the first data from the new laboratory."
More information: Benjamin Kostiuk et al, Type VI secretion system mutations reduced competitive fitness of classical Vibrio cholerae biotype, Nature Communications (2021). DOI: 10.1038/s41467-021-26847-y Journal information: Nature Communications




Comment: See also: