The fat sand rat is a strange creature. It lives in burrows, eats around 80 per cent of its body mass in leaves each day and doesn't drink water. But the really odd thing about this gerbil is that some of its DNA appears to be missing.
No doubt you have heard of dark matter, which is thought to make up over a quarter of the universe. We know it's there; we just haven't been able to detect it. Well, something similar is afoot in the genome. My colleagues and I have dubbed this elusive genetic matter "dark DNA". And our investigations into the sand rat are starting to reveal its nature.
The discovery of dark DNA is so recent that we are still trying to work out how widespread it is and whether it benefits those species that possess it. However, its very existence raises some fundamental questions about genetics and evolution. We may need to look again at how adaptation occurs at the molecular level. Controversially, dark DNA might even be a driving force of evolution.
The sand rat (Psammomys obesus) is a desert species native to North Africa and the Middle East, but put it in a lab and something strange happens. When fed a "normal" diet - the standard fare for laboratory rodents - sand rats tend to become obese and develop type 2 diabetes. This was discovered in the 1960s, and has made sand rats the focus of study for biologists interested in understanding nutrition-induced diabetes in humans. Yet, in all that time, the mystery of why these gerbils are so susceptible to the disease has remained unsolved.
My main work interest is venomous snakes. Admittedly, the link with sand rats is tenuous - at best, the rodents might be seen as dinner for the snakes - so the species is not an obvious target of study for me. But I have always relished a mystery, and the more I found out about sand rats, the more intrigued I became. The smoking gun seemed to be a gene called Pdx1. The Pdx1 protein it codes for has many roles, including in the development of the pancreas and in switching the insulin gene on and off.
Being crucial for normal physiology, this gene is found in all vertebrates. Intriguingly, though, genetic studies had failed to spot it in sand rats. Yet they have a normal pancreas and are able to secrete insulin. That didn't make sense. What was really going on here?
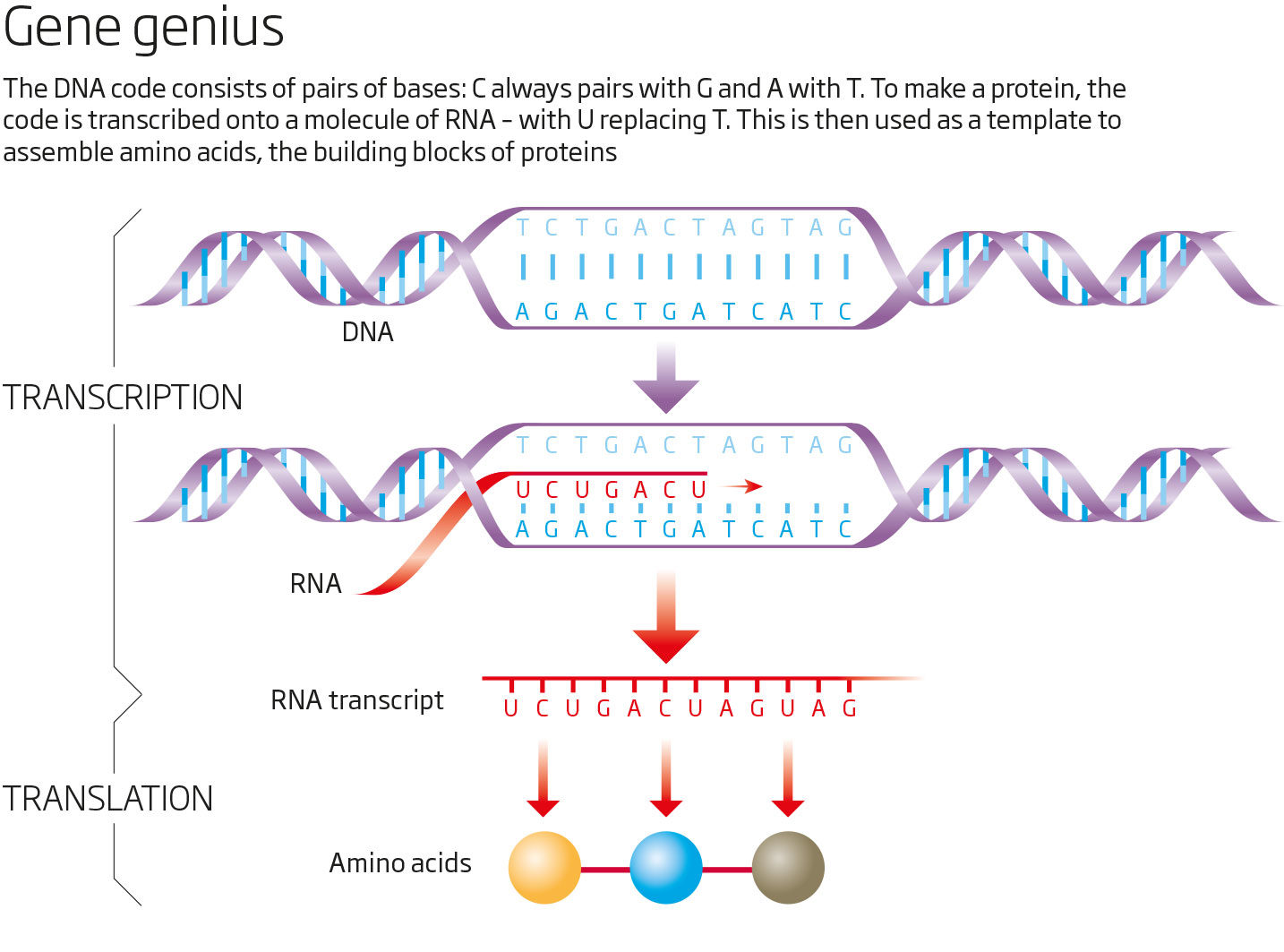
Luckily for me, my fascination with this paradox coincided with a revolution in genetics that made it possible to investigate. I teamed up with 17 researchers at nine institutions around the world, including the Beijing Genomics Institute, and we set out to sequence the entire sand rat genome. What we discovered was even more baffling; Pdx1 was not the only missing gene. In fact, a big chunk of DNA containing nearly 90 genes, which are found on the same chromosome in other animals, was nowhere to be seen. Many of these genes, like Pdx1, are essential for survival. What's more, we found their corresponding RNA transcripts - copies of stretches of genetic code that cells use as templates to make proteins (see diagram). But where were the genes?
The big clue came when we scrutinised the RNA transcripts. The genetic code is comprised of four bases, A, T, G and C. What made the sequences in these transcripts so bizarre was the very high level of two of these, G and C. None of us had ever seen anything like it. But we realised it might explain why the corresponding DNA appeared to be missing - standard sequencing technology is not very good at picking up sections of DNA with high levels of G and C. So we set out to reveal the elusive DNA in a different way: using caesium chloride ultracentrifugation. This involves spinning chopped-up DNA in a highly concentrated salt solution very fast - at least 40,000 revolutions per minute - for three days so that denser fragments, like those rich in GC bases, sink to the bottom. Having separated this out, we attempted to sequence it alone.
It worked. What we found was a mutation hotspot - a region of DNA with an extraordinarily large number of mutations, many of them changes from A or T to G or C bases. Sand rat Pdx1, for example, contains more mutations than any other version of the gene we know of in the animal kingdom - resulting in a Pdx1 protein that, in just one key region that binds to DNA, has at least 15 amino acids differing from the normal version.
It is extremely rare for vertebrates to have any mutations in this region. Mutations usually compromise the function of a gene, and the genes in our chunk of hard-to-detect dark DNA are so essential for survival that they have barely changed over the course of evolution. Yet somehow the sand rat's Pdx1 gene, along with others here, are managing to function despite the dramatic mutation levels. This discovery has forced us to revise our ideas about how much change a gene can tolerate and still work.
The extreme divergence of Pdx1 might help explain why sand rats develop diabetes, if their Pdx1 protein turns out to be not as effective as its counterpart in other animals. It also explains why Pdx1 initially appeared to be absent. But in solving the mystery of the missing DNA, we have raised an intriguing possibility. We know that standard genome sequencing has trouble picking up sections of DNA containing lots of G and C bases, so perhaps sand rats are not alone in carrying these sorts of mutation hotspots. Dark DNA might be lurking in other genomes.
In fact, 12 other species of gerbils apparently lack Pdx1, suggesting that they too may possess dark DNA. We are now looking into that. What's more, a striking parallel to the sand rat story is found in birds. The many bird genomes sequenced so far seemed to lack more than 270 genes present in most other vertebrate genomes, including important genes such as the one coding for leptin, a hormone that regulates hunger.
However, new research by Fidel Botero-Castro of Ludwig-Maximilian University in Munich, Germany, and colleagues reveals that birds do make RNA transcripts of these "missing" genes. What's more, the sequences are very high in G and C bases. Sound familiar? In fact, the researchers estimate that around 15 per cent of all bird genes have been overlooked in previous studies.
This hints that dark DNA could be quite widespread. If so, we may have to rethink some current ideas about how genomes evolve. By comparing the thousands of whole genomes that have been sequenced in the past decade (see "Deciphering the code of life"), biologists are trying to work out which genes have been lost in certain lineages and which new ones have arisen. This helps them see what makes groups of organisms different from one another and how adaptation occurs at the molecular level.
If dark DNA is common, that would throw a spanner in the works because genes we thought were missing might actually be present. It may be time to take another look at the genomes sequenced to date to find out whether we have got the full picture. At the least, we should be alert to the possibility of dark DNA when sequencing new genomes.

Even more intriguing is how dark DNA might influence evolution. Most textbooks describe evolution as a two-step process. First, a steady trickle of random genetic mutation creates variation in an organism's DNA. Then, natural selection acts like a filter, deciding which mutations are passed on. This usually depends on whether they confer some sort of advantage, although not everything produced over the course of evolution is an adaptation. So, natural selection is the sole driving force pushing the direction in which organisms evolve. But add dark DNA to the picture, and that's not necessarily the case. If genes contained within these mutation hotspots have a greater chance of mutating than those elsewhere, they will display more variation on which natural selection can act, so the traits they confer will evolve faster. In other words, dark DNA could influence the direction of evolution, giving a driving role to mutation. Indeed, my colleagues and I have suggested that mutation rates in dark DNA may be so rapid that natural selection cannot act fast enough to remove deleterious variants in the usual way. Such genes might even become adaptive later on, if a species faces a new environmental challenge.
Dual control?
The idea of mutation-driven evolution is controversial, but it is not without precedent. Since the mid-1970s, esteemed molecular biologist Masatoshi Nei has argued that the most important driving force behind evolution occurs at the molecular level, in the variation created within DNA by mutation. Without this spontaneous variation, natural selection would have nothing to work with, making it of secondary importance. The discovery of dark DNA lends weight to this way of thinking. Of course, it's not a straight choice between mutation and natural selection. In the sand rat, for example, a massively elevated rate of mutation in the many dark DNA genes could have had a significant effect upon the species' evolutionary trajectory. Nevertheless, some selection must also act upon these genes, otherwise mutation would run rampant, creating a region of nonsense with no functional genes, and the species would not have survived.
In truth, it is difficult to determine whether the sand rat has benefited overall from its mutation hotspot. You would expect its extreme mutation to be a problem, otherwise why would proteins such as Pdx1 be virtually identical in all other animals? But the sand rat's dark DNA could have led to some adaptations that would not have arisen under normal circumstances. Perhaps these have allowed it to survive on a diet of very nutritionally poor food with little access to water, and so thrive in a harsh desert environment with very few competitors. On the other hand, if sand rats eat nutritionally rich food they will develop diabetes and die. That could mean they are constrained to living in deserts. So dark DNA could be both their liberator and their jailer.
In general, the implications of dark DNA remain enigmatic. One thing is for sure, though. It shows we still have a lot to learn about how genomes evolve at the molecular level, and how these processes give rise to the awe-inspiring diversity of life on Earth.
Deciphering the code of life
It took over a decade, a huge consortium and an estimated $2.7 billion to sequence the human genome. Since the project was completed in 2003, there have been huge advances in technology, making sequencing much faster and cheaper. To date, the full genomes of some 15,000 species have been deciphered. And that's just the start - the race to commercialise whole genome sequencing is so intense that it may not be long before we routinely decipher the entire genetic blueprint of individuals, including fetuses in the womb.
Genetic sequencing involves deducing the exact order of the four nucleotides or bases - known as A, T, G and C - paired along a DNA strand. Pioneering technologies, such as the one devised by Frederick Sanger in the 1970s, were largely manual. Sanger sequencing entails using a single strand of DNA as a template to "grow" a complementary strand, one base at a time in a test tube, using special markers that are then read off to give the sequence. It is accurate but extremely time-consuming; a skilled worker might decipher 10,000 base pairs on a good day. The human genome consists of some 3.2 billion base pairs.
These days, sequencing is largely automated, so the process is much faster. Multiple copies of a DNA strand are first chopped up at random into small fragments - usually between 100 and 150 bases long - which are then sequenced individually before being pieced back together by computer programs that match overlapping sections. But there's a problem. This "next-generation" sequencing is not very good at deciphering stretches of DNA dominated by just two bases, such as G and C, because this makes it hard to reassemble overlapping fragments. As a result, we may have overlooked substantial chunks of DNA in the genomes sequenced to date. My colleagues and I have dubbed this "dark DNA".
Newer sequencing methods are more accurate. They can process stretches of DNA up to several thousand bases long, reducing the problem of deciphering overlapping areas. With the technology rapidly improving, dark DNA will come into view. We may even discover new surprises in genomes we thought we had decoded.




Nice to see a scientist exhibiting a bit of humility or caution for once