These atlases, published on 19 July in Nature1-3, are examples of a powerful and increasingly popular approach to studying the organs of the body in both health and disease. Each comprises hundreds of thousands of data points about gene activity and protein production in individual cells, which are then mapped to their specific location in the organ.
The hope is that the atlases will eventually yield clues about how to diagnose and treat disorders that can arise when those cells become injured or dysfunctional. "These cells organize themselves into neighbourhoods, towns, countries," says Michael Snyder, a geneticist at Stanford University in California and an author of the study looking at the intestine3. "And it affects their function."
Cell atlases
Technologies that allow researchers to monitor gene activity in individual cells have helped to spur the production of many cell atlases in recent years, including maps of blood vessels in the brain4 and of various types of tumour5-7. With time, these technologies have become more sophisticated, allowing researchers to incorporate information about a cell's location and interrogate gene activity more thoroughly.
The latest papers take this further by evaluating the abundance of dozens of proteins in each cell. The research is part of a consortium called the Human Biomolecular Atlas Program (HuBMAP), which is funded by the US National Institutes of Health and aims to develop tools to map out the cells of the human body.
In one study, researchers incorporated a twist. Rather than mapping tissues in a single organ, they studied the interface between two: the placenta and the uterus1. The team used data from 500,000 cells and 588 uterine arteries to learn about how cells from the fetus invade and remodel blood vessels in the lining of the uterus so that they become larger and better able to deliver nutrients in the later stages of pregnancy. "They invade the arteries and replace the maternal cells, which is wild," says Michael Angelo, a pathologist at Stanford University and an author of the study.
Errors in this process have been associated with pre-eclampsia and other conditions that can endanger the health of both fetus and mother.
Angelo's results mesh well with data published earlier this year that catalogued gene activity in cells that form the placenta8, says Roser Vento-Tormo, a geneticist at the Wellcome Sanger Institute in Cambridge, UK. In particular, data on the expression of 37 proteins in each cell give researchers a high-resolution view of the communication between fetal and maternal cells, she says. "If we want to look at disease, we need to understand in depth what happens in healthy conditions."
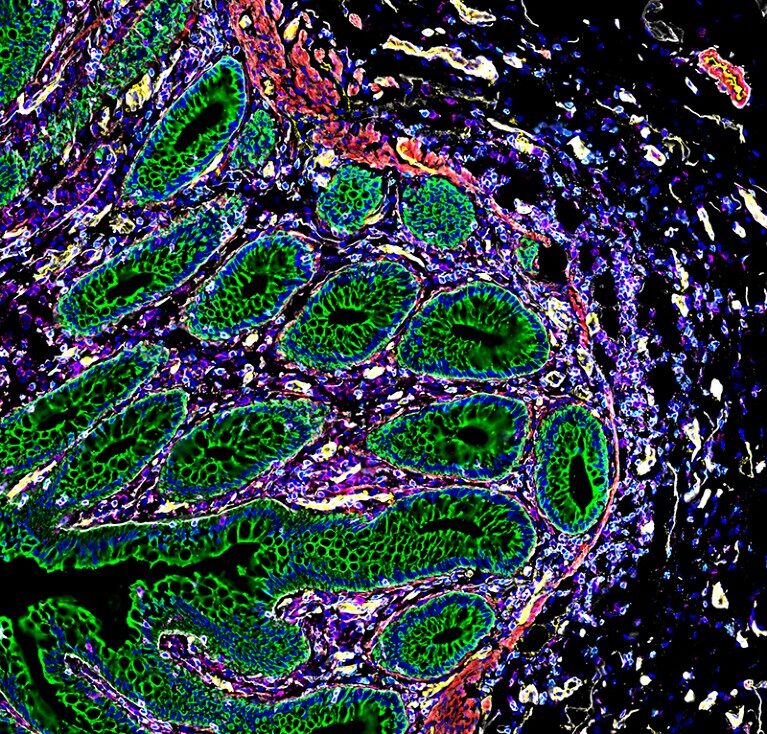
Intestinal neighbourhoods
One of the latest studies compares healthy and injured kidney cells2. "We were able to build pathways of how the cells might be travelling from healthy to injured, and the rest stops along the way," says co-author Sanjay Jain, a pathologist at Washington University School of Medicine in St Louis, Missouri.
And one study evaluates cells taken from eight sites along the intestine and finds a number of what Snyder calls neighbourhoods of cells with unique characteristics3.
The beauty of the studies lies not just in their direct findings, but also in how they have integrated vast reams of data, says Vento-Tormo. Researchers are also working to boost the diversity of tissue donors to cell-atlas projects, she says, and to develop ways of doing expanding from 2D to 3D analyses.
Future studies will no doubt look at more tissues, disease states and developmental stages, says Sarah Teichmann, a geneticist at the Sanger Institute who participates in HuBMAP. "There are a lot of other tissues and organs in the body," she says. "There are exciting opportunities ahead."
References
- Greenbaum, S. et al. Nature 619, 595-605 (2023).Article Google Scholar
- Lake, B. B. et al. Nature 619, 585-594 (2023).Article Google Scholar
- Hickey, J. W. et al. Nature 619, 572-584 (2023).Article Google Scholar
- Sun, N. et al. Nature Neurosci. 26, 970-982 (2023).Article PubMed Google Scholar
- Zhang, M. et al. Preprint at biorXiv https://doi.org/10.1101/2023.03.06.531348 (2023).
- Wu, S. Z. et al. Nature Genet. 53, 1334-1347 (2021).Article PubMed Google Scholar
- Ozato, Y. et al. Cell Rep. 42, 111929 (2023).Article PubMed Google Scholar
- Arutyunyan, A. et al. Nature 616, 143-151 (2023).Article PubMed Google Scholar



Med system is up the shoot.
I doubt this sort of treatment is for our benefit, beyond us plebs. For the specials only so they can live forever because where would the world be without them right?