This paper provides a tempting explanation for the otherwise unexpected findings that the spike protein mRNA lasts at least two months and that the spike protein itself lasts at least four months after mRNA vaccination.
This paper did not show that this happens in live humans, that it happens in healthy liver cells, or that it is integrated into the chromosomal DNA such that it would reliably persist after cell division and be evenly split among newly divided cells. It especially did not show that it is written into the germline, such that the children of vaccinated parents would contain it in their own, brand new genomes.
The quality and import of this paper are mediocre. The authors could have looked for integration into the chromosomes, but didn't. They attributed the reverse transcription to a human enzyme known as LINE-1; they could have demonstrated this clearly by suppressing the production of that enzyme, but didn't. My main problem with this paper, though, is that their description of how they isolated the genomic DNA and purified it from any contaminating mRNA is incomplete.1 I am waiting to hear back from them by email for some clarification on this. Pending satisfactory answers to my questions, I tentatively consider this the first evidence that the vaccine spike protein mRNA can be reverse transcribed into DNA within human cells.
Despite these limitations to the paper, all these questions should have been settled with well done experiments prior to ever having conducted human trials, not researched sloppily as an afterthought while we were halfway through vaccinating the entire world.
Previous Evidence on Integration of the Viral Genome into the Human Genome
We should view the findings of this paper in the context of the existing controversy over whether the genome of the natural virus can be incorporated into human chromosomes.
In May of 2021, a paper was published in the Proceedings of the National Academy of Sciences arguing that the viral genome of SARS-CoV-2, the virus that causes COVID, is both reverse transcribed and integrated into the chromosomes of human cells.
They were trying to solve the puzzle of why some COVID patients who strictly quarantined were PCR positive for months despite not generating any culturable virus that could spread and despite having no way of being reinfected.
Some of these patients also showed indirect evidence of reverse transcripotion. Unlike DNA, which is made of two mirror-opposite strands bound together (positive and negative), the RNA from SARS-CoV-2 is single-stranded (only positive). When the virus replicates, virtually all the RNA is identical (the positive strand). If the viral RNA were to be reverse transcribed into DNA, the DNA would become double-stranded with two mirror opposites (positive and negative). A few patients who were inexplicably PCR-positive for long periods of time had "mirror opposite" (negative) strand RNA present at orders of magnitude higher fractions than would happen with the virus replicating as normal. The authors considered this a signature suggestive of reverse transcription.
Classically, reverse transcription of RNA to DNA and integration into the host genome is thought to be the province of specific types of viruses known as retroviruses. HIV is an example. However, we have our own reverse transcription machinery. Presumably, its healthful function is DNA repair, but it also allows the possibility of retrotranscribing viruses.
The authors of the May 2021 paper noted that other viruses that are not retroviruses, such as vesicular stomatitis virus or lymphocytic choriomeningitis virus have already been shown to be reverse transcribed and integrated into the human chromosomes with human enzymes.
This paper looked at the nucleocapsid gene, not the spike gene. The nucleocapsid is the protein that coats the virus. Since they didn't look at the spike protein, we can't generalize from the natural spike to the vaccine spike. However, if the nucleocapsid integrates, it shouldn't be considered wildly out of expectation for the spike to integrate, whether from the natural virus or the vaccine. And since other viruses have already been shown to be reverse transcribed and integrated by human enzymes, nothing is particularly implausible about it happening with SARS-CoV-2.
On technical grounds, unfortunately, these findings were disputed back and forth, with numerous papers accumulating by October, 2021 arguing the finding may have been spurious, but with no clear resolution.
This is More Concerning for the Vaccine Spike Than the Natural Spike
If we suppose that both the natural and vaccine spike are capable of reverse transcription and integration, we should be much more concerned about the vaccine spike for the vast majority of people.
This is because the natural virus for most people will be limited to the respiratory tract, and to a lesser extent the eyes or the gut, where mucosal immunity will prevent it from systemically circulating to all of our internal organs. For example, virus is only found in the blood of 44% of those on a ventilator, 27% of those hospitalized, and 13% of those treated as outpatients. As I pointed out in Explaining the Hospitalization Paradox, "Almost certainly the incidence is even lower in those with mild cases who never seek hospital treatment, and it is almost certainly non-existent in those who were exposed without ever feeling ill."
By contrast, the vaccine spike is guaranteed systemic circulation by being injected into the arm and bypassing the mucosal immune system.
The mRNA of the spike protein is highly modified to make it evade the innate immune system, last for much longer, and be translated into protein at a much higher rate. Thus, vaccination not only guarantees systemic exposure to spike protein, in radical contrast to natural infection, but also makes it likely that the magnitude of that exposure will be very high compared to what would occur in the minority of natural infections that involve systemic exposure.
Integration Is Not Necessary for Gene Expression
Zooming out a bit, one of the criticisms of the February 25 paper is that it did not show the reverse transcribed DNA was integrated into the chromosomes. This is an important point, but it is also critical to note that integration is not required for gene expression.
It has been known since the 1980s that retroviruses produce a substantial amount of DNA that forms independent circles within the nucleus rather than integrating into the chromosomes. Decades of research showed that this circular extra-chromosomal retroviral DNA was indeed expressed, forming mRNA that was then translated into protein. While circular extra-chromosomal DNA is more short-lived and expressed at a lower rate than integrated, chromosomal DNA, it can persist for months in some cell types and is expressed at a sufficient rate to change the phenotype of the cells. In the case of HIV, for example, non-integrated DNA is capable of lowering CD4 expression in T cells.
Retroviral DNA is circularized by human enzymes rather than viral enzymes, so the circularization doesn't depend on the particular virus. Even free linear DNA is expressed, just at an even lower rate than circular DNA.
More recently, extra-chromosomal circular DNA was shown to be present in large amounts in healthy human cells. Little is known about how its expression profile compares to that of chromosomal DNA, but we know that some of it is expressed. This is because the two ends of DNA that join together to form a circle form a unique junction point that is not present when the same genes are found as part of the chromosomes. mRNA transcripts that were formed by reading the DNA across this junction point are found within the cell, showing that at least some of these DNA circles are in fact transcribed.
The role of extrachromosomal circular DNA in healthy cells is not well characterized. However, it may play a role in enhancing the copy number of highly expressed genes so their expression can be amplified. More highly expressed genes are also more likely to accumulate mutations, so perhaps extrachromosomal copies of those genes allow a reservoir of information that can be used to repair the genes when mutations prove deleterious.
The combined length of the extrachromosomal DNA in human cells is equivalent to 12.6% of the length of the chromosomal DNA, suggesting that it should be seen as a substantial part of the human genome. Its expression profile is poorly understood, but it is better understood than much of the "junk DNA" that is considered part of the genome. It may be more dynamic and less stable than the chromosomal DNA, but this also is not well understood. Most of the circles are not attached to centromeres, which mediate even division of chromosomes when a cell divides, so the circles most likely do not separate into dividing cells evenly. All of this just makes extrachromosomal DNA look like a more fluid component of the genome than the chromosomal DNA, but part of the genome nonetheless.
Did the Feb 25 Paper Show the Spike Protein Is "Written Into the Genome"?
I do not believe that integration into chromosomes should be required to say something is "written into the genome." However, I do believe that the fate of the reverse-transcribed DNA would have to be more clearly characterized than was achieved in the February 25 paper. Therefore, I believe my early posts on social media that the paper showed it was "written into the human genome" went too far.
However, there is no evidence that it does not integrate into the chromosomes yet, so I would not rule this out.
Putting integration aside, though, I believe a likely alternative fate of the DNA is to circularize inside the nucleus, and if it does this, is expressed, and remains stable through cell division I would personally consider that being written into the genome.
If it becomes extrachromosomal DNA, and is expressed, that would allow continued production of mRNA that would, in turn, allow continued reverse transcription of DNA. Even if the circular DNA is less stable than chromosomal DNA, it could be continually replaced. The spike protein mRNA could attain a permanent or quasi-permanent status within any cell lineage in which it had become established.
Presumably, it would stay within the cell to which it was originally delivered. In dividing cells, if it persists to cell division, it would presumably persist in the divided cells, just not dividing perfectly equally like the chromosomes ideally do. However, it would at least be conceivable that it could leave those cells in exosomes, which would allow for an even more concerning circulation throughout the body that could resemble a systemic infection.
All of this is conceivable without integration into the chromosomes, but integration cannot yet be ruled out.
Why Does the Spike Protein mRNA Last At Least Two Months and the Spike Protein At Least Four Months?
I would like to turn our attention in a different direction: Something has to be invoked to explain the unexpectedly long duration of the spike protein and its mRNA after vaccination.
In addition to the late-January paper showing the spike protein and its mRNA persist for 60 days after vaccination in the lymph nodes of the arm pits, Andreas Oehler brought my attention to a mid-November paper showing that spike protein is found in circulating sacs known as exosomes four months after vaccination.
While the exosome finding could theoretically be attributed to spike protein being preserved from degradation for up to four months (this would be a bit odd given that the point of vaccination is to direct an immune response against it), the lymph node paper unambiguously shows the mRNA continues to be found at least two months out.
Given the basic science we know up to this point, should we have expected the mRNA to last for two months?
Let's begin by looking at what we should expect for a half-life of the mRNA. This is the time it takes for half of it to be degraded.
Remember that if the mRNA is not self-replicating because there is no virus in the vaccine, because it is not reverse transcribed, because it is not integrated into the human genome, and because it is not packaged with any kind of self-replicating machinery, then we are simply starting with the mRNA contained in the vaccine while it disappears over time according to its half-life.
Here is what we know:
The median half-life of mRNA in human cells depends on the cell type and study method but is generally between three to ten hours. Around ~7.5% of mRNAs have half-lives greater than 24 hours. Some known examples include the globin genes needed for hemoglobin synthesis, whose mRNAs can have half-lives as high as 29 hours. The half-life for tyrosine hydroxylase mRNA increases from 10 hours to 30 hours under conditions of oxygen deprivation. One paper didn't single out the longest-lasting mRNA, but the longest-lasting family of enzymes were in glutamine metabolism, with an average half-life of 4.8 days.
Previous measures of coronavirus mRNAs have yielded half-lives of several hours.
The vaccine spike mRNA is highly modified to last longer than the natural spike mRNA. I was not able to find a direct measurement of its half-life, but I did find a theoretical model suggesting that under physiological conditions it would be about 2.4 days.
We can use this model's 2.4-day half-life as our expectation, and the longest-known half-life reported in human cells of 4.8 days as our upper bound of plausibility.
As a general rule of thumb, something is 97% gone after five half lives, 99% gone in 6-7 half lives, and 99.9% gone after ten half lives. We should therefore expect the spike mRNA to be gone over 12-20 days and should consider it stretching the bounds of plausibility to last more than 48 days.
So how is the mRNA turning up easily detectable in the lymph nodes of the armpits 60 days after vaccination?
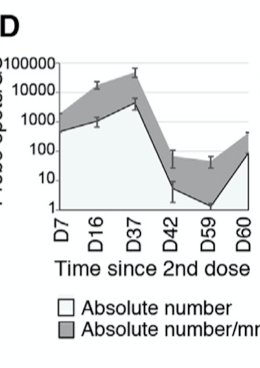
The late-January lymph node paper does not allow us to track the decay of mRNA molecules over time, but it does provide one semi-quantitative measurement of mRNA concentration at different time points after vaccination cross-sectionally:
Different measurements in the graph represent different patients who had a lymph node biopsy at different lengths of time after vaccination. Since different time points represent different people — we call this "cross-sectional" data — there will be a lot more noise in the data than if everyone had measurements at every time point and we could watch how the mRNA concentration changed over time in each person.
The way they are measuring this is by bathing a biopsy sample in a solution with a vaccine-specific probe and seeing how much of the probe sticks. This doesn't tell us the actual concentration of the mRNA, but it should at least give us a reasonable sense of the relative differences in concentration at different time points. Thus, it is "semi-quantitative" rather than "quantitative." This also makes the data noisier.
The bottom line represents the absolute number of probes that stuck, while the top line represents the number of probes that stuck per square millimeter of tissue measured.
Within the top line, the values increase from 2000 at day 7 to 44,730 at day 37, bottom out at 45 on day 59, and rise back to 400 on day 60.
As described in my second footnote,2 explaining the persistence of the mRNA at the 60-day timepoint at the levels shown here would require invoking an implausibly long half-life of 9-26 days.
This is 2 to 6 times the upper bound of plausibility using the longest-lasting human mRNAs, 4 to 11 times the half-life predicted by the theoretical model, and 22-108 times the half-life that would be expected of typical human mRNAs or natural coronavirus mRNAs.
Since this is semi-quantitative, cross-sectional data, we must regard any of these back-of-the-envelope calculations with a grain of salt. The point is not to try to precisely quantify the half-life, but just to get a general feel for whether it makes sense that the mRNA is hanging around this long.
It doesn't.
Not without invoking some unforeseen mechanisms of preserving it or replicating it.
While the lipid nanoparticles were expected (page 54) to take 4-5 months to be 95% gone, they should be quickly delivering their mRNA cargo to cells, otherwise the immune response would take forever to get going. Perhaps they have some slow-drip component to keep delivering this cargo over the course of those 4-5 months?
Perhaps lymph nodes have some unforeseen way of dramatically extending the half-life of mRNA? The point is we must invoke something unexpected to explain the persistence over time.
While the topic of reverse transcription is not settled, it should now be fully investigated as a leading candidate to explain the puzzling persistence of spike mRNA for at least two months and of spike protein for at least four months.
If the mRNA has some way of replicating, this could explain the apparent 22-fold increase from 2000 to 44,730 from day 7 to day 37 in the lymph node paper.
This could explain why there is nothing remotely resembling an idealized exponential decay graph, which should slope continuously downward with an initially steep slope that decelerates over time.
This could explain why just when it threatens to bottom out at the end of the graph, it rebounds.
The Bottom Line
Here's the bottom line:
- Pending satisfactory answers to my questions for the authors (footnote 1), I tentatively regard the February 25 paper as the first evidence showing the vaccine spike protein mRNA is reverse transcribed into DNA within human cells.
- While my original characterization of the paper on social media as showing the spike mRNA was "written to the human genome" went too far, I do not believe that integration into the chromosomes needs to be shown to justify this characterization. Rather, the organization of the DNA within the nucleus needs to be clarified (whether it is in the nucleus or elsewhere; if in the nucleus, whether it chromosomal or extrachromosomal; if the latter, whether it is circular or linear), and if it turns out to be nuclear, relatively stable, and expressed, I would consider it "written to the human genome."
- This finding needs to be replicated by other groups in other types of cells to develop consensus around the finding and to show that the phenomenon is not peculiar to this cell line. However, it does not at all invalidate the paper for them to have chosen a cell line that would make the finding easier. This is a completely rational way to illustrate proof of principle that something can happen. Generalizing to other cell types is the next step.
- This should be studied in animal experiments to characterize which tissues, if any, are vulnerable to this in vivo, how often it occurs, and what its consequences are.
- Human studies should look for spike mRNA-derived DNA in the months after mRNA vaccination in chromosomal, extrachromosomal circular, or extrachromosomal linear forms, keeping in mind that sampling error could be a big problem if this phenomenon occurs in a patchy and inconsistent manner.
- Reverse transcription should be considered as a leading candidate to explain why the spike mRNA lasts for at least two months and the spike protein itself lasts for at least four months after vaccination. These long durations do not make sense without invoking some unexpected mechanisms of preserving or replicating the mRNA.
- While this may have not been expected, and while it still is not settled, this was known as plausible the entire time and this should have been settled with well-done experiments before the human trials were conducted. It is public health malfeasance that we are left to argue about medicore-quality studies on this topic while we are halfway through vaccinating the world.
Footnotes
- They link to a protocol for preparing mouse tails to be genotyped, but the protocol does not explain its principle or describe how it would isolate genomic DNA. I would have expected this to be paired with alcohol precipitation and centrifugation, which are not described. They also do not state whether the RNase they used from Qiagen to purify the isolated genomic DNA from any contaminating RNA was RNase A or H (Qiagen sells both) and I believe they should have stated this clearly and also justified why they wouldn't expect the pseudouridine modification in the mRNA to interfere with it (some RNA modifications interfere with digestion from certain RNases). The most convincing part of Figure 5 is that control lane 5, representing RNA isolated from Huh7 treated with 0.5 mcg/mL BNT162b2 after six hours, does not contain any positive staining, unlike the genomic DNA isolated from cells treated similarly.If the positive staining for the genomic DNA sample were due to contamination with RNA, then the RNA sample should be even more positive, but it is negative. However, I find that difficult to reconcile to Figure 2 V1 at the 6h time point, which shows abundant mRNA and yet seems to be the identical treatment as Figure 5 Control 5. Perhaps Figure 5 Control 5 represents an RNA extraction from the isolated genomic DNA; if so, the figures make sense, but they do not state this. It appears they are missing steps in their methods section and have some lack of clarity in the labeling of their figures. I am waiting to hear back for clarification by email.
- From Figure 7D. The graph is logarithmic, so each tick on the y axis is ten times the tick preceding it. I used the rulers in PowerPoint to measure the distance between ticks, converted this to a decimal between 0 and 1, raised ten to the power of this decimal, then multiplied it by the tick the value had surpassed, which gives the approximate value being shown. For example, a value that is 65% of the way between 100 and 1000 would represent (10^0.65)*100=447. Half-lives were calculated with this calculator. Within the top line, the values increase from 2000 at day 7 to 44,730 at day 37, bottom out at 45 on day 59, and rise back to 400 on day 60. If we impute the day 7 data to day 0, we have a decrease from 2000 to 400 over 60 days.This implies a wildly implausible half-life of 26 days. If we generously impute the day 37 value to day 0, we have a decrease from 44,730 to 400 over 60 days, which still gives an implausibly long half-life of almost 9 days.The only way we could put the half-life anywhere within an expected range would be to start at day 37, where a decrease from 44,730 to 400 over 23 days gives a more reasonable half-life of 3.4 days. We could be even more generous and narrow our view from day 37 to day 59, getting a decrease from 44,730 to 45 over 22 days, which would bring the half-life to 2.2 days. In fact, the most generous thing we could do is look at the lower line with the absolute counts and trace it from day 37 to day 59, where it goes from 4,473 to 1.2, bringing the half-life down to 1.9 days. But all of these generous ways to reduce the apparent half-life down to something normal require ignoring the first month!
If we simply try to account for the 60-day stay, we have to invoke a half-life somewhere between 9 and 26 days, all far longer than could have been predicted and outside the bound of plausibility set by the longest-known half-lives in human cells.A further note: while the 22-fold increase from day 7 to day 37 looks suspiciously like the mRNA is replicating itself, there are other possibilities.Apart from the noise introduced by the cross-sectional and semi-quantitative natures of the data, the mRNA might be progressively accumulating in the lymph nodes, transported from elsewhere, as part of the immune response. However, invoking this explanation requires assuming that the mRNA is being exported from cells that it initially entered, which opens a whole other set of concerns. For example, if the mRNA can be replicated and exported, this would start to resemble a systemic infection.




I wonder what they will come up with on the protection brainstorm.