A new study adds to an emerging, radically new picture of how bacterial cells continually repair faulty sections of their DNA.
Published online May 16 in the journal Cell, the report describes the molecular mechanism behind a DNA repair pathway that counters the mistaken inclusion of a certain type of molecular building block, ribonucleotides, into genetic codes. Such mistakes are frequent in code-copying process in bacteria and other organisms.
Given that ribonucleotide misincorporation can result in detrimental DNA code changes (mutations) and DNA breaks, all organisms have evolved to have a DNA repair pathway called ribonucleotide excision repair (RER) that quickly fixes such errors.
Last year a team led by Evgeny A. Nudler, PhD, the Julie Wilson Anderson Professor of Biochemistry in the Department of Biochemistry and Molecular Pharmacology at NYU Langone Health, published two analyses of DNA repair in living E. coli cells. They found that most of the repair of certain types of DNA damage (bulky lesions), such as those caused by UV irradiation, can occur because damaged code sections have first been identified by a protein machine called RNA polymerase. RNA polymerase motors down the DNA chain, reading the code of DNA "letters" as it transcribes instructions into RNA molecules, which then direct protein building.
Dr. Nudler and coworkers found that during this transcription process, RNA polymerase also finds DNA lesions, and then serves as a platform for the assembly of a DNA repair machine called nucleotide excision repair (NER) complex. NER then snips out faulty DNA found and replaces it with an accurate copy. Without the action of RNA polymerase, little NER, if any, occurs in living bacteria.
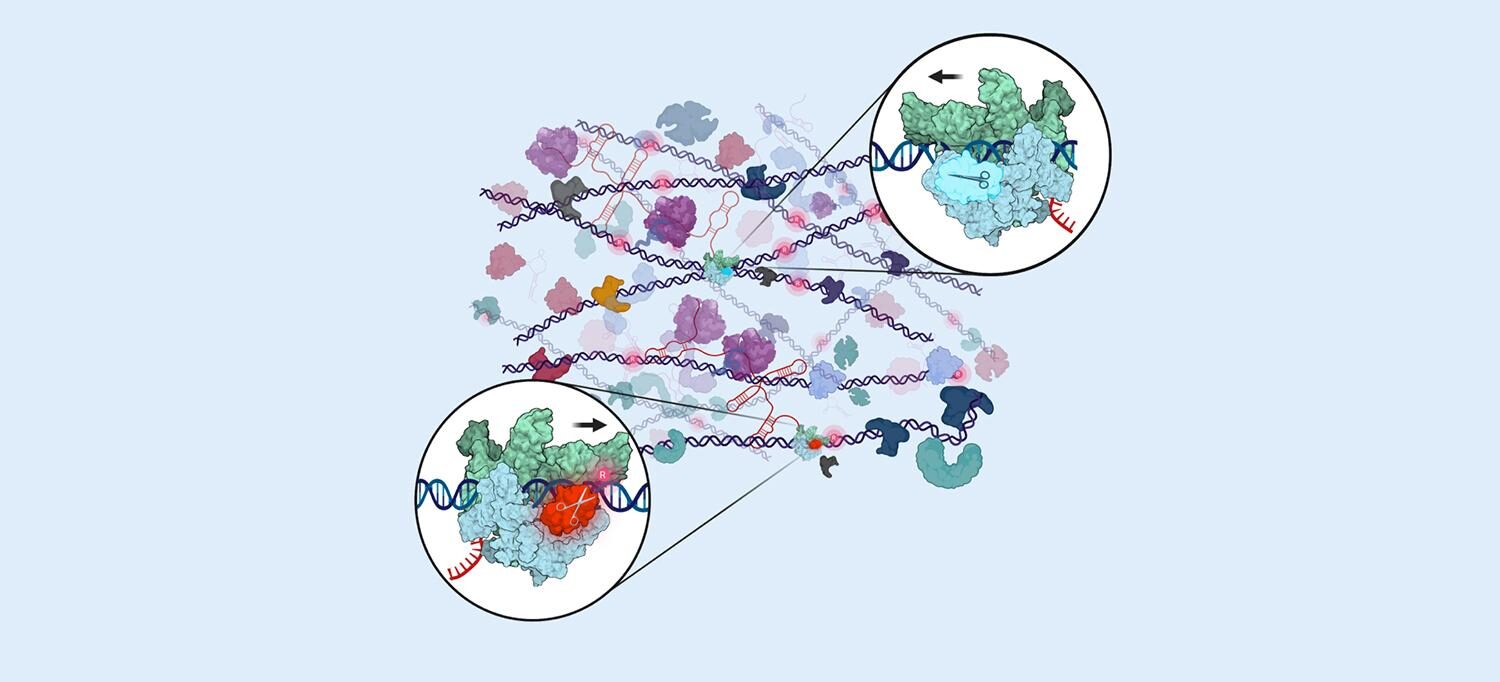
Now the new study in Cell provides the first evidence that, like in the NER pathway, RER is tightly coupled to transcription. The study authors found evidence that the key enzyme involved in RER, RNaseHII, also cooperates with RNA polymerase as it scans for misincorporated ribonucleotides in the DNA chains of living bacterial cells.
"Our results continue to inspire a rethinking of certain basic principles in the DNA repair field," says Dr. Nudler, also an investigator with the Howard Hughes Medical Institute. "Moving forward, our team plans to investigate whether RNA polymerase scans DNA for all kinds of problems and triggers repair genome-wide, not only in bacteria, but in human cells as well."
Cutting-Edge Techniques
Ribonucleotides (the building blocks of RNA) and deoxyribonucleotides (DNA components) are related compounds. As cells copy and build DNA chains in bacterial cells, they often mistakenly incorporate ribonucleotides into DNA chains in place of deoxyribonucleotides because they differ by only a single oxygen atom, say the study authors. In bacterial cells, DNA polymerase III is known to make about 2,000 of these mistakes every time it copies a cell's genetic material. To maintain genome integrity, the bulk of misplaced ribonucleotides are removed by the RER pathway, but a key question had been about how RNaseHII finds relatively rare ribonucleotide lesions amidst an "ocean" of intact cellular DNA codes so quickly.
As they did in their 2022 studies, the researchers used quantitative mass spectrometry and in vivo protein-protein crosslinking to map the distances between chemically linked proteins, and so determined the key surfaces of RNaseHII and RNA polymerase as they interact in living bacterial cells. In this way they determined that most RNaseHII molecules couple with RNA polymerase.
In addition, they used cryogenic electron microscopy (CryoEM) to capture the high-resolution structures of RNaseHII bound to RNA polymerase to reveal the protein-protein interactions that define the RER complex. Further, structure-guided genetic experiments that weakened the RNA polymerase/RNaseHII interaction compromised RER.
"This work supports a model where RNaseHII scans DNA for misplaced ribonucleotides by riding on RNA polymerase while it moves along DNA," says first study author Zhitai Hao, a postdoctoral scholar in Dr. Nudler's lab. "This work is vital for our basic understanding of the DNA repair process and has far-reaching clinical implications."
Along with Dr. Nudler and Hao, study authors in the Department of Biochemistry and Molecular Pharmacology at NYU Grossman School of Medicine were Manjunath Gowder, Binod Bharati, Vitaly Epshtein, Vladimir Svetlov, and Ilya Shamovsky. The study was supported by the National Institute of Health, the Howard Hughes Medical Institute, and by the Blavatnik Family Foundation.
Media Inquiries
Greg Williams
Phone: 212-404-3500
gregory.williams@nyulangone.org



Comment: See also: