Poore et al. used The Cancer Genome Atlas (TCGA) — an online resource that includes DNA and RNA sequences — to analyse data for 33 cancer types, totalling more than 17,000 samples from some 10,000 patients. They analysed data sets derived from bulk tumour samples (primary tumours as well as recurrent ones, and tumours that had spread through metastasis), normal adjacent tissue and blood samples. The authors used multiple computational approaches, including independently trained artificial-intelligence (AI) models, to filter, normalize and classify microbial sequences in these samples. After stringent filtering approaches to address potential contamination and other variables, the group classified 7.2% of the total sequencing reads as non-human. Approximately one-third of those mapped to known sequences of bacterial, archaeal or viral origin, and 12.6% of these resolved to a particular genus from one of these groups.
The authors next trained machine-learning models to use these sequences to distinguish between cancer types and between different stages of the same cancer type, as well as between tumours and normal tissue. Overall, the models performed well in discriminating between cancer types and between cancer and normal tissue, but showed some variability in their ability to discriminate between various stages of cancer.
The researchers also tested the biological relevance of the microbial profiles against known microbial associations with cancer. In line with previous reports, they found Fusobacterium in gastrointestinal tumours, and viruses such as Alphapapillomavirus and Hepacivirus in cervical cancer, head and neck cancer and hepatocellular cancer.
Poore et al. next explored microbial signatures in the blood of people with cancer, using AI models that analysed whole-genome sequences from the TCGA cohort of individuals. Their findings suggest that blood-borne microbial DNA (mbDNA) could be used to discriminate between cancer types. The group sought to validate its mbDNA models against existing cell-free tumour DNA (ctDNA) assays in a separate cohort, which included 69 individuals without cancer and 100 who had prostate cancer, lung cancer or a skin cancer called melanoma (Fig. 1). The authors' models were generally good at discriminating between cancer types, although there were some limitations. Further validation of these results is needed in larger cohorts across cancer types using dedicated methods.
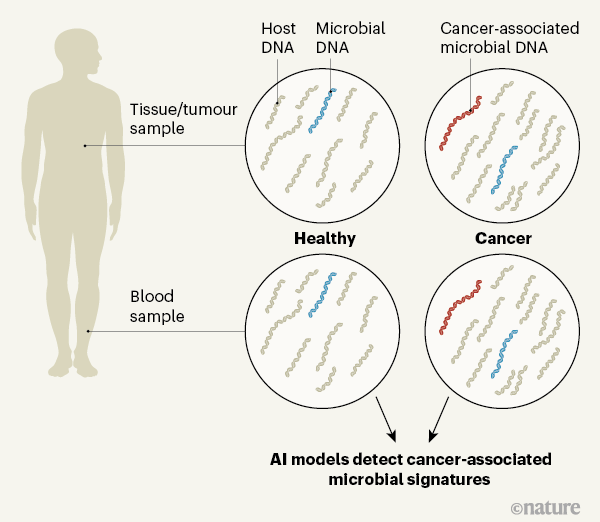
One limitation is that TCGA samples were not collected in a manner that controlled for contamination by microbes or mbDNA. This contamination could have been introduced at any time between sample collection and sequencing. Poore et al. and others6 tried to control for this through stringent filtering of potential contaminants; however, such approaches might limit our ability to detect the full complement of microbes present in tumours. In addition, DNA and RNA sequencing for human studies might not be performed in a way that enables microbes to be characterized completely. Future studies that build on the current work should involve analysis of carefully curated tissues and blood using appropriate sequencing techniques to allow for characterization of microbial signatures.
In addition to validating the presence of these microbes in tumours and blood in cancer, it will be important to gain insights into their distribution and function. Poore et al. and others6 identified microbial signatures in tumours on the basis of nucleic-acid sequences; however it is not known where these microbes are located (within or around tumour cells, immune cells or in connective tissue known as the stroma) and whether or not they are alive. And more work will be needed to determine whether the microbes are driving cancer or are merely passengers in an altered tumour microenvironment. There are clear examples of how microbes in tumours might contribute to cancer development and to resistance to cancer therapy3,10. However, other data suggest that the presence of microbes in tumours is associated with better long-term outcomes11.
Finally, further mechanistic insights into how microbes enter and persist in cancerous tissue are needed, as well as research into how best to target them for treatment and even cancer prevention. Such strategies will need to be nuanced, and must take into account the potential effect on all microbial niches, because many of the body's resident microbes have a crucial role in overall physiology. Although some preclinical studies suggest that co-targeting microbes and tumour cells with antibiotics and chemotherapy is associated with delayed tumour outgrowth10,12, other work suggests that treatment with broad-spectrum antibiotics13 can worsen the outcomes of people receiving immunotherapy, probably owing to disruption of the gut microbiome. Thus, there is context dependence, which must be taken into consideration. Nonetheless, the opportunities for both clinical advances and basic insights that are presented by an ability to monitor and manipulate the microbiome are tantalizing.
doi: 10.1038/d41586-020-00637-w
References
1. Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. Nature Med. 25, 377-388 (2019).
2. Dejea, C. M. et al. Proc. Natl Acad. Sci. USA 111, 18321-18326 (2014).
3. Kostic, A. D. et al. Cell Host Microbe 14, 207-215 (2013).
4. Marshall, B. J. & Warren, J. R. Lancet 323, 1311-1315 (1984).
5. Bullman, S. et al. Science 358, 1443-1448 (2017).
6. Robinson, K. M., Crabtree, J., Mattick, J. S. A., Anderson, K. E. & Dunning Hotopp, J. C. Microbiome 5, 9 (2017).
7. Nakatsuji, T. et al. Nature Commun. 4, 1431 (2013).
8. Whittle, E., Leonard, M. O., Harrison, R., Gant, T. W. & Tonge, D. P. Front. Microbiol. 9, 3266 (2018).
9. Poore, G. D. et al. Nature https://doi.org/10.1038/s41586-020-2095-1 (2020).
10. Geller, L. T. et al. Science 357, 1156-1160 (2017).
11. Riquelme, E. et al. Cell 178, 795-806 (2019).
12. Sethi, V. et al. Gastroenterology 155, 33-37 (2018).
13. Wilson, B. E., Routy, B., Nagrial, A. & Chin, V. T. Cancer Immunol. Immunother. 69, 343-354 (2020).



I'm taking the next interstellar ferry back to the Pleiades.