For that reason, DNA can be used to encode messages:
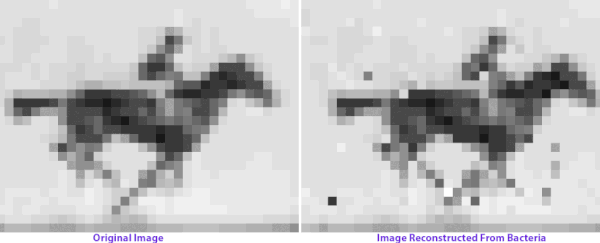
If just encoding text, one way is to convert each letter of the alphabet into a three-letter code. Using three bases, such as A, C, and T, gives 27 combinations - enough for the English alphabet plus a space - with a code such as AAA = A, AAC = B, and so on (1 in graphic below). However, researchers often want to encode more than just text, so most current methods instead first translate data into binary code - the language of 1s and 0s used in electronic media. Using binary, the four bases of DNA could theoretically store up to two bits of information per nucleotide, with a code such as A = 00, C = 01, and so on. --CATHERINE OFFORD, "INFOGRAPHIC: WRITING WITH DNA" AT THE SCIENTISTIn 2017, one Harvard group encoded a video, an image of one of the earliest surviving motion pictures, in a DNA sample from bacteria:
But in some ways, our genomes are much more powerful than words. They are part of a process that utters not just ideas but living beings. Including human beings, who ourselves have ideas.
In August 2017, researchers announced that they had used DNA to encode malware to hack a computer program that reads genetic sequences:
In new research they plan to present at the USENIX Security conference on Thursday, a group of researchers from the University of Washington has shown for the first time that it's possible to encode malicious software into physical strands of DNA, so that when a gene sequencer analyzes it the resulting data becomes a program that corrupts gene-sequencing software and takes control of the underlying computer. While that attack is far from practical for any real spy or criminal, it's one the researchers argue could become more likely over time, as DNA sequencing becomes more commonplace, powerful, and performed by third-party services on sensitive computer systems. --ANDY GREENBERG, "BIOHACKERS ENCODED MALWARE IN A STRAND OF DNA" AT WIREDThe researcher/hackers merely wanted to demonstrate the possibility, in an age when DNA is becoming popular culture:
Between startups like 23andMe, makers of an at-home saliva-based DNA kit that promises to help users learn more about their health and family history, and Embark Veterinary, which helps pet owners and breeders learn about ancestry and disease risk of dogs through saliva swabs, DNA testing is having a bit of a moment. "Security Researchers Inject DNA with Malware - But Don't Panic Yet" at Data Center KnowledgeWhat the researchers did was to write a piece of attack software that, 37% of the time, survived translation from physical DNA to FASTQ, a digital storage format for DNA sequences and then could get into the computer's memory and start running whatever it was coded to do.
Now, they did make things easier for themselves in that they deliberately inserted a flaw in the open source code of the compression program, fqzcomp, to be sure they had something to attack. However, they weren't exactly cheating because they surveyed commonly used DNA sequencing software and found three genuine vulnerabilities.
So yes, it's still science fiction - for now. Like all languages, the language that forms us can be misused and we must anticipate the challenge.
[1] Here's the Significance statement of the 2019 paper:
Genomes appear similar to natural language texts, and protein domains can be treated as analogs of words. To investigate the linguistic properties of genomes further, we calculated the complexity of the "protein languages" in all major branches of life and identified a nearly universal value of information gain associated with the transition from a random domain arrangement to the current protein domain architecture. An exploration of the evolutionary relationship of the protein languages identified the domain combinations that discriminate between the major branches of cellular life. We conclude that there exists a "quasi-universal grammar" of protein domains and that the nearly constant information gain we identified corresponds to the minimal complexity required to maintain a functional cell. --LIJIA YU, DEEPAK KUMAR TANWAR, EMANUEL DIEGO S. PENHA, YURI I. WOLF, EUGENE V. KOONIN, AND MALAY KUMAR BASU, ""GRAMMAR OF PROTEIN DOMAIN ARCHITECTURES"" AT PNASSee also: How a computer programmer looks at DNA And finds it to be "amazing" code



Comment: But if you ask hardcore neo-Darwinists like PZ Myers, DNA isn't really a code. It just looks like a code, acts like a code, and can be used as a code. But really, it's not a code. Because that would imply a coder.