© University of California, San Francisco
Researchers from the
University of California, San Francisco have combined advanced computer technology, DNA-sequencing technology and biochemical techniques to create a new method of observing how cells convert DNA into RNA, and ultimately discovered what makes genes turn on and off.
Jonathan Weissman, Ph.D., study leader and a professor in the UCSF Department of Cellular and Molecular Pharmacology, and L. Stirling Churchman, Ph.D., of the UCSF Department of Cellular and Molecular Pharmacology as well as the California Institute for Quantitative Biosciences, have developed a new method of understanding the information held in an organism's genome in order to figure out how
cells read DNA to create RNA as well as turn on and off.
"The genome is the hard drive of the cell," said Churchman. "Until now, we've been able to see the information that the hard drive contains as well as see the result after the cell has read that information, but we didn't know which precise data it was accessing. Here, we've been able to see which data it is accessing, with a high enough resolution to also be able to see how it's actually working."
Up until this point, scientists believed that less than 5 percent of the human genome is interpreted and turned into RNA. But recent advancements in not only this study, but others as well, have confirmed that a majority of the DNA is transcribed.
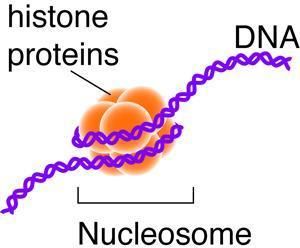
To study this process, Weissman and Churchman used the genome of baker's yeast because previous studies have already mapped out the genome for this organism. For instance, previous studies have located the positions of nucleosomes, which are structures created by strands of DNA that wrap around histone proteins, throughout the genome of the baker's yeast. Histone proteins have markings that show whether a gene should be turned on or off while also showing a history of what has occurred in that section of the gene code. In addition, histone proteins are capable of offering an idea of what should occur in the future.
By adding their own maps of RNA production to the maps made of the genome of the baker's yeast, Weissman and Churchman were able to watch the process directly. They witnessed polymerase coming into contact with the histone proteins during transcription. The histone markings controlled the "junk RNA" being produced from DNA, and as the polymerase enzyme scooted along the genome transcribing DNA into RNA, nucleosomes acted as speed bumps.
"There is a long history of trying to look at how genes are turned on," said Weissman "So far, nothing has been analogous at this resolution and depth."
This study was published in
Nature.

Reader Comments
to our Newsletter